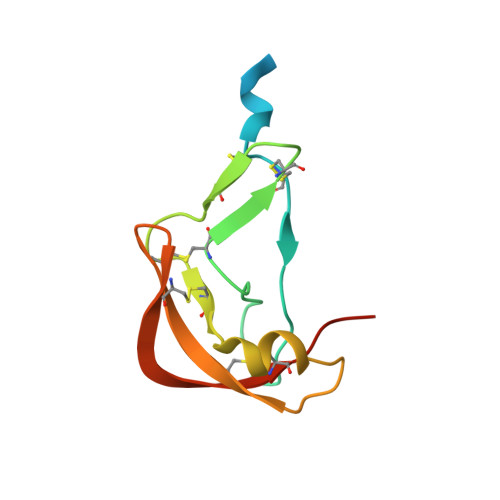
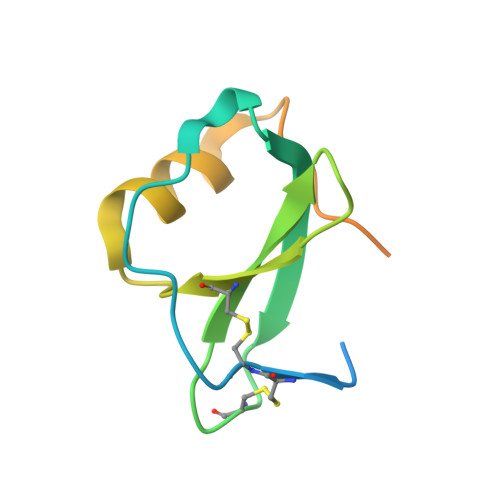
Structural basis of chemokine recognition by the class A3 tick evasin EVA-ACA1001.
Devkota, S.R., Aryal, P., Wilce, M.C.J., Payne, R.J., Stone, M.J., Bhusal, R.P.(2024) Protein Sci 33: e4999-e4999
- PubMed: 38723106
- DOI: https://doi.org/10.1002/pro.4999
- Primary Citation of Related Structures:
8FK9 - PubMed Abstract:
Ticks produce chemokine-binding proteins, known as evasins, in their saliva to subvert the host's immune response. Evasins bind to chemokines and thereby inhibit the activation of their cognate chemokine receptors, thus suppressing leukocyte recruitment and inflammation. We recently described subclass A3 evasins, which, like other class A evasins, exclusively target CC chemokines but appear to use a different binding site architecture to control target selectivity among CC chemokines. We now describe the structural basis of chemokine recognition by the class A3 evasin EVA-ACA1001. EVA-ACA1001 binds to almost all human CC chemokines and inhibits receptor activation. Truncation mutants of EVA-ACA1001 showed that, unlike class A1 evasins, both the N- and C-termini of EVA-ACA1001 play minimal roles in chemokine binding. To understand the structural basis of its broad chemokine recognition, we determined the crystal structure of EVA-ACA1001 in complex with the human chemokine CCL16. EVA-ACA1001 forms backbone-backbone interactions with the CC motif of CCL16, a conserved feature of all class A evasin-chemokine complexes. A hydrophobic pocket in EVA-ACA1001, formed by several aromatic side chains and the unique disulfide bond of class A3 evasins, accommodates the residue immediately following the CC motif (the "CC + 1 residue") of CCL16. This interaction is shared with EVA-AAM1001, the only other class A3 evasins characterized to date, suggesting it may represent a common mechanism that accounts for the broad recognition of CC chemokines by class A3 evasins.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Monash Biomedicine Discovery Institute, Monash University, Clayton, VIC, Australia.