Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
Caroli, J., Mattevi, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Lysine-specific histone demethylase 1A | 871 | Homo sapiens | Mutation(s): 0 Gene Names: KDM1A, AOF2, KDM1, KIAA0601, LSD1 EC: 1.14.99.66 | 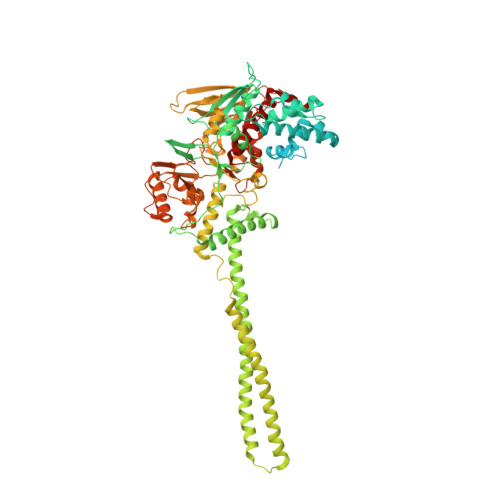 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O60341 (Homo sapiens) Explore O60341 Go to UniProtKB: O60341 | |||||
PHAROS: O60341 GTEx: ENSG00000004487 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O60341 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| REST corepressor 1 | 144 | Homo sapiens | Mutation(s): 0 Gene Names: RCOR1, KIAA0071, RCOR | 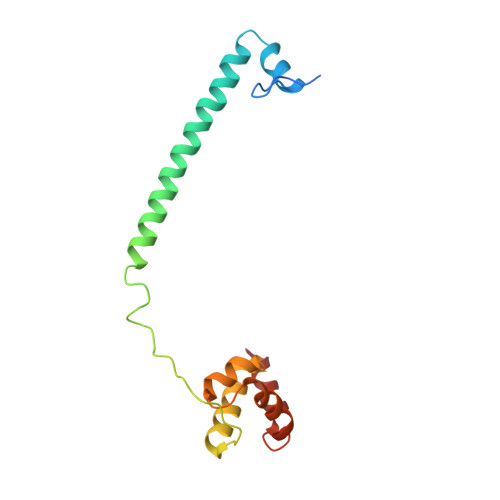 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9UKL0 (Homo sapiens) Explore Q9UKL0 Go to UniProtKB: Q9UKL0 | |||||
PHAROS: Q9UKL0 GTEx: ENSG00000089902 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9UKL0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| XZQ (Subject of Investigation/LOI) Query on XZQ | F [auth A] | [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(1R,3R,3aS,13R)-1-hydroxy-10,11-dimethyl-4,6-dioxo-3-[3-(phenylcarbamoyl)phenyl]-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]pentyl dihydrogen diphosphate C43 H48 N10 O17 P2 UUPSWUMMRQTZIA-QZCXPCBOSA-N |  | ||
| XZU (Subject of Investigation/LOI) Query on XZU | C [auth A], D [auth A], E [auth A] | 3-[(1R,2S)-2-(cyclobutylamino)cyclopropyl]-N-phenylbenzamide C20 H22 N2 O OGPXXRHRKMJPBF-MOPGFXCFSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 118.346 | α = 90 |
| b = 179.743 | β = 90 |
| c = 232.329 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Aimless | data scaling |
| PHENIX | refinement |
| XDS | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Italian Association for Cancer Research | Italy | IG19808 |