Potent Long-Acting Inhibitors Targeting the HIV-1 Capsid Based on a Versatile Quinazolin-4-one Scaffold.
Gillis, E.P., Parcella, K., Bowsher, M., Cook, J.H., Iwuagwu, C., Naidu, B.N., Patel, M., Peese, K., Huang, H., Valera, L., Wang, C., Kieltyka, K., Parker, D.D., Simmermacher, J., Arnoult, E., Nolte, R.T., Wang, L., Bender, J.A., Frennesson, D.B., Saulnier, M., Wang, A.X., Meanwell, N.A., Belema, M., Hanumegowda, U., Jenkins, S., Krystal, M., Kadow, J.F., Cockett, M., Fridell, R.(2023) J Med Chem 66: 1941-1954
- PubMed: 36719971
- DOI: https://doi.org/10.1021/acs.jmedchem.2c01732
- Primary Citation of Related Structures:
8FIU - PubMed Abstract:
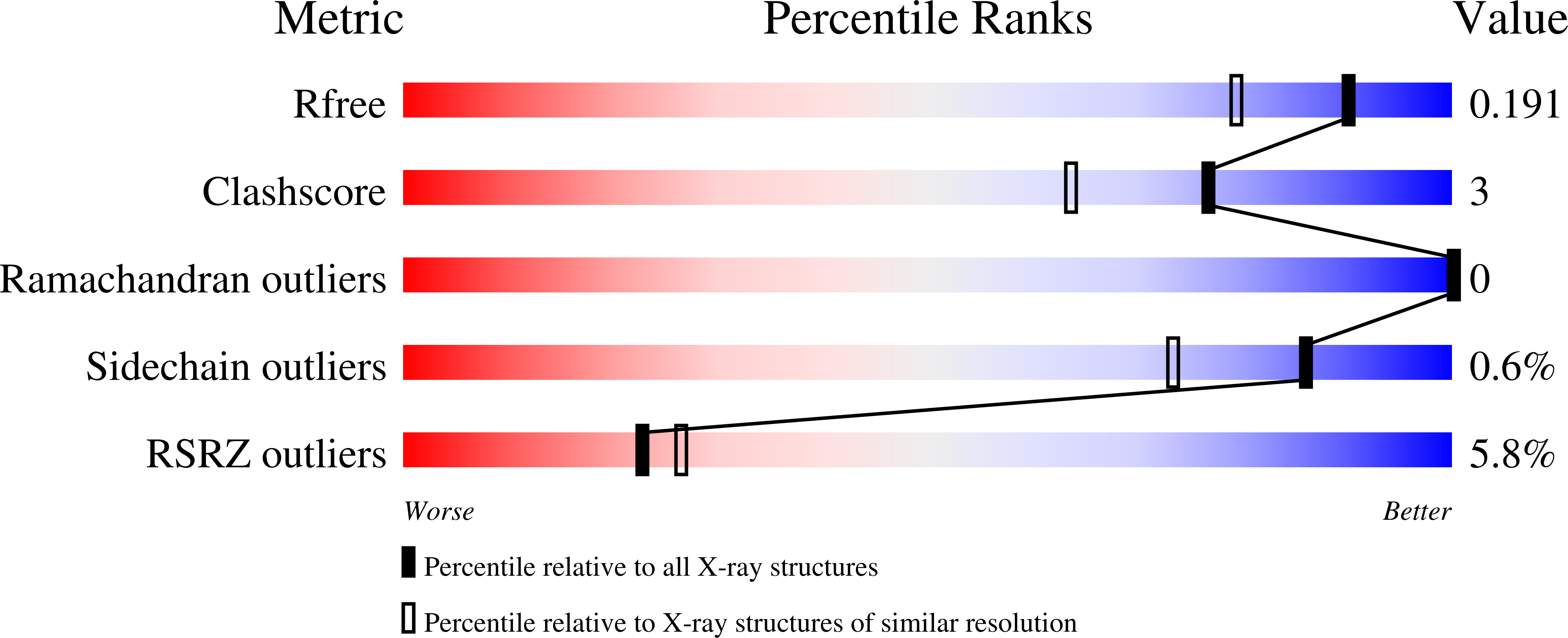
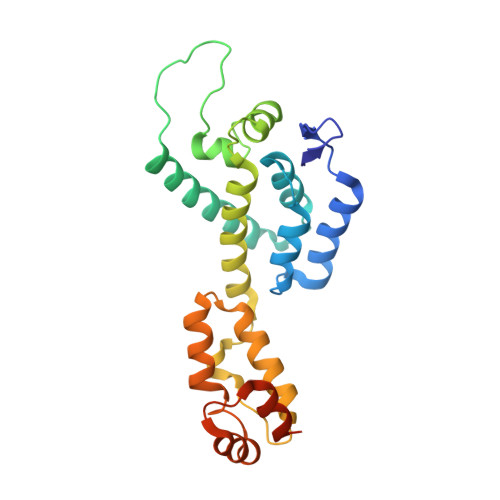
Long-acting (LA) human immunodeficiency virus-1 (HIV-1) antiretroviral therapy characterized by a ≥1 month dosing interval offers significant advantages over daily oral therapy. However, the criteria for compounds that enter clinical development are high. Exceptional potency and low plasma clearance are required to meet dose size requirements; excellent chemical stability and/or crystalline form stability is required to meet formulation requirements, and new antivirals in HIV-1 therapy need to be largely free of side effects and drug-drug interactions. In view of these challenges, the discovery that capsid inhibitors comprising a quinazolinone core tolerate a wide range of structural modifications while maintaining picomolar potency against HIV-1 infection in vitro , are assembled efficiently in a multi-component reaction, and can be isolated in a stereochemically pure form is reported herein. The detailed characterization of a prototypical compound, GSK878, is presented, including an X-ray co-crystal structure and subcutaneous and intramuscular pharmacokinetic data in rats and dogs.
Organizational Affiliation:
Discovery Chemistry, ViiV Healthcare, Branford, Connecticut 06405, United States.