Potent, Selective, and Membrane Permeable 2-Amino-4-Substituted Pyridine-Based Neuronal Nitric Oxide Synthase Inhibitors.
Vasu, D., Do, H.T., Li, H., Hardy, C.D., Awasthi, A., Poulos, T.L., Silverman, R.B.(2023) J Med Chem 66: 9934-9953
- PubMed: 37433128
- DOI: https://doi.org/10.1021/acs.jmedchem.3c00782
- Primary Citation of Related Structures:
8FG9, 8FGA, 8FGB, 8FGC, 8FGD, 8FGE, 8FGF, 8FGG, 8FGH, 8FGI, 8FGJ, 8FGK, 8FGL, 8FGM, 8FGN, 8FGO, 8FGP, 8FGQ, 8FGR, 8FGS, 8FGT, 8FGU, 8FGV - PubMed Abstract:
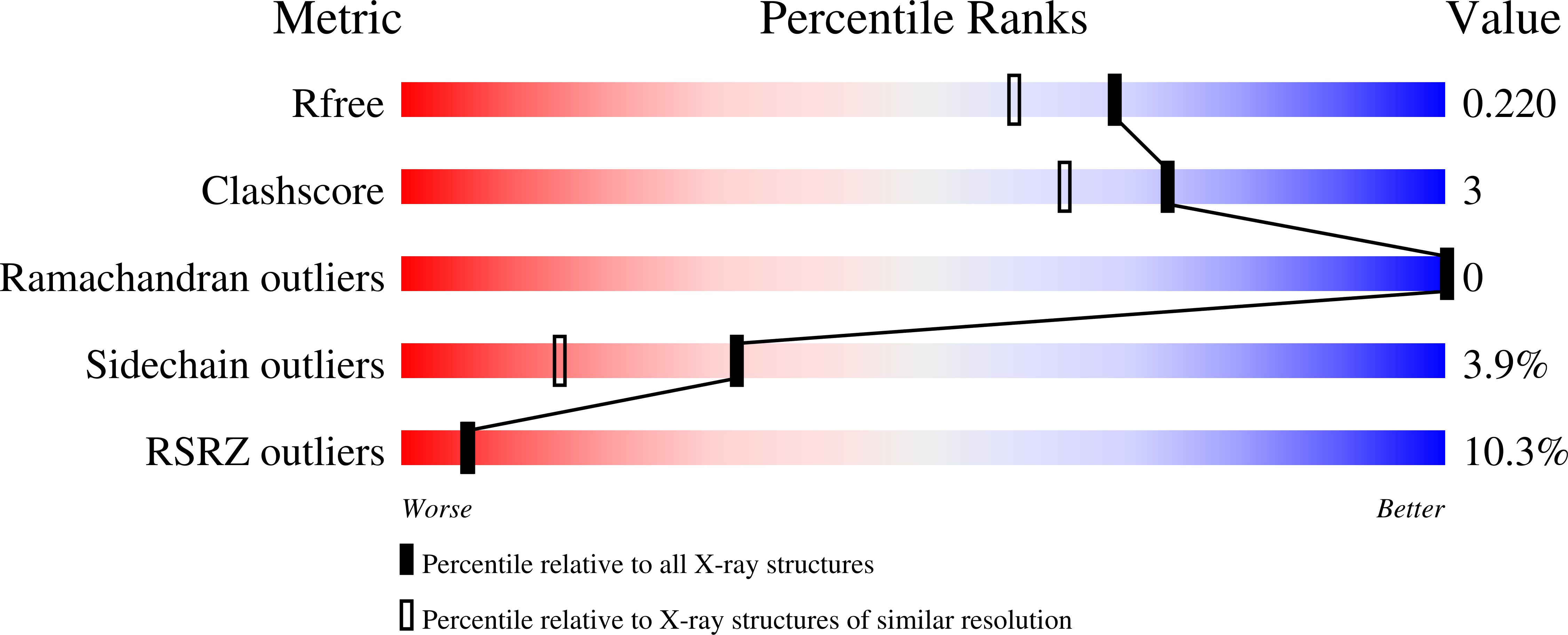
A series of potent, selective, and highly permeable human neuronal nitric oxide synthase inhibitors (hnNOS), based on a difluorobenzene ring linked to a 2-aminopyridine scaffold with different functionalities at the 4-position, is reported. In our efforts to develop novel nNOS inhibitors for the treatment of neurodegenerative diseases, we discovered 17 , which showed excellent potency toward both rat ( K i 15 nM) and human nNOS ( K i 19 nM), with 1075-fold selectivity over human eNOS and 115-fold selectivity over human iNOS. 17 also showed excellent permeability ( P e = 13.7 × 10 -6 cm s -1 ), a low efflux ratio (ER 0.48), along with good metabolic stability in mouse and human liver microsomes, with half-lives of 29 and >60 min, respectively. X-ray cocrystal structures of inhibitors bound with three NOS enzymes, namely, rat nNOS, human nNOS, and human eNOS, revealed detailed structure-activity relationships for the observed potency, selectivity, and permeability properties of the inhibitors.
Organizational Affiliation:
Department of Chemistry, Department of Molecular Biosciences, Chemistry of Life Processes Institute, Center for Developmental Therapeutics, Northwestern University, 2145 Sheridan Road, Evanston, Illinois 60208-3113, United States.