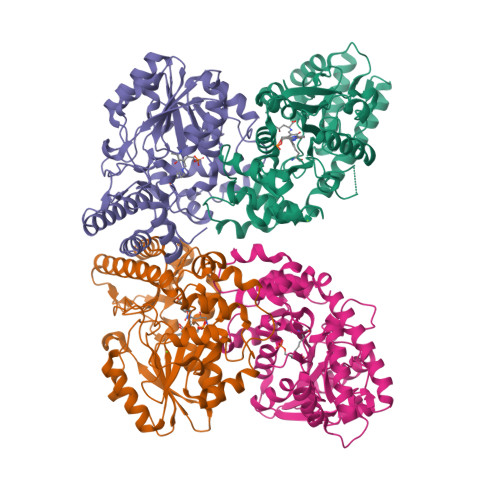
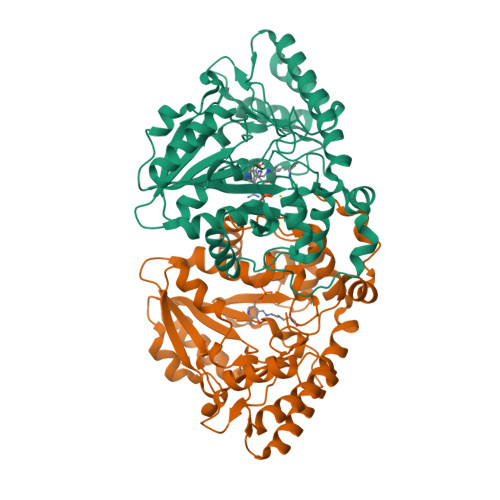
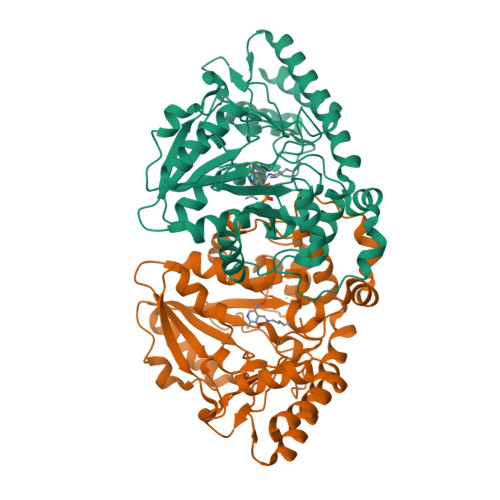
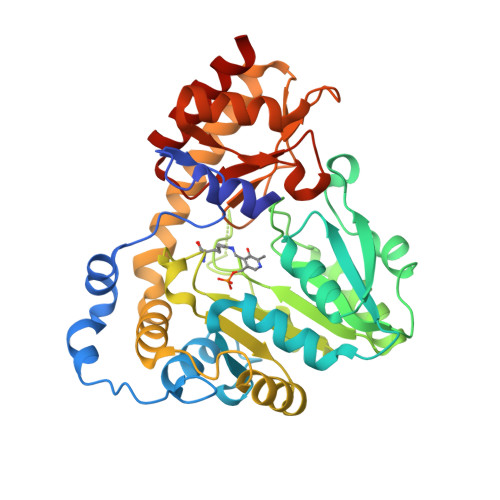
Mechanistic and Structural Insights into a Divergent PLP-Dependent l-Enduracididine Cyclase from a Toxic Cyanobacterium.
Cordoza, J.L., Chen, P.Y., Blaustein, L.R., Lima, S.T., Fiore, M.F., Chekan, J.R., Moore, B.S., McKinnie, S.M.K.(2023) ACS Catal 13: 9817-9828
- PubMed: 37497377
- DOI: https://doi.org/10.1021/acscatal.3c01294
- Primary Citation of Related Structures:
8FFT, 8FFU - PubMed Abstract:
Cyclic arginine noncanonical amino acids (ncAAs) are found in several actinobacterial peptide natural products with therapeutically useful antibacterial properties. The preparation of ncAAs like enduracididine and capreomycidine currently takes multiple biosynthetic or chemosynthetic steps, thus limiting the commercial availability and applicability of these cyclic guanidine-containing amino acids. We recently discovered and characterized the biosynthetic pathway of guanitoxin, a potent freshwater cyanobacterial neurotoxin, that contains an arginine-derived cyclic guanidine phosphate within its highly polar structure. The ncAA l-enduracididine is an early intermediate in guanitoxin biosynthesis and is produced by GntC, a unique pyridoxal-5'-phosphate (PLP)-dependent enzyme. GntC catalyzes a cyclodehydration from a stereoselectively γ-hydroxylated l-arginine precursor via a reaction that functionally and mechanistically diverges from previously established actinobacterial cyclic arginine ncAA pathways. Herein, we interrogate l-enduracididine biosynthesis from the cyanobacterium Sphaerospermopsis torques-reginae ITEP-024 using spectroscopy, stable isotope labeling techniques, and X-ray crystallography structure-guided site-directed mutagenesis. GntC initially facilitates the reversible deprotonations of the α- and β-positions of its substrate before catalyzing an irreversible diastereoselective dehydration and subsequent intramolecular cyclization. The comparison of holo - and substrate-bound GntC structures and activity assays on site-specific mutants further identified amino acid residues that contribute to the overall catalytic mechanism. These interdisciplinary efforts at structurally and functionally characterizing GntC enable an improved understanding of how nature divergently produces cyclic arginine ncAAs and generate additional tools for their biocatalytic production and downstream biological applications.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, Santa Cruz, California 95064, United States.