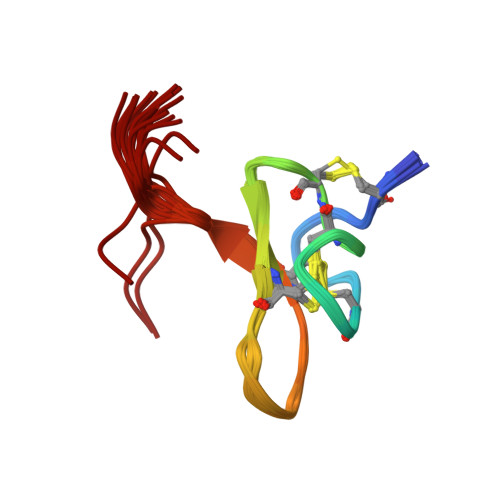
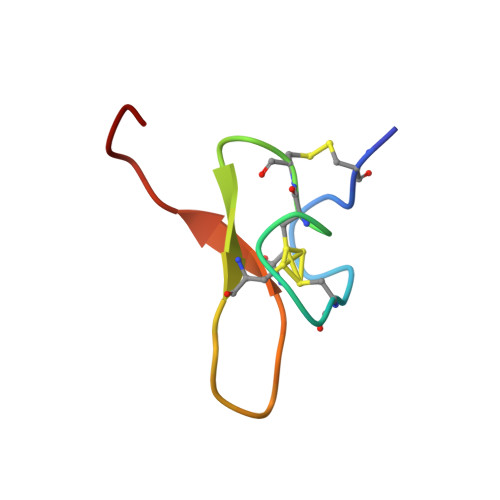
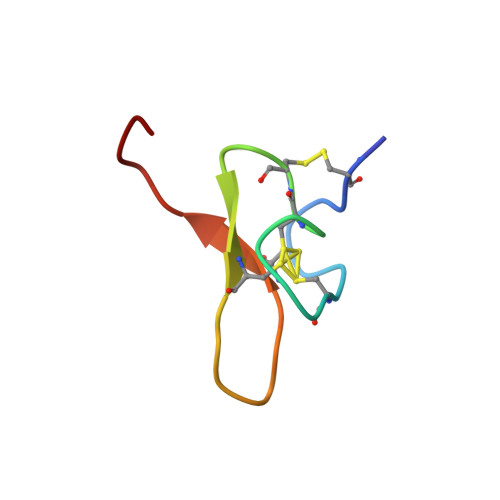
Pmu1a, a novel spider toxin with dual inhibitory activity at pain targets hNa V 1.7 and hCa V 3 voltage-gated channels.
Giribaldi, J., Chemin, J., Tuifua, M., Deuis, J.R., Mary, R., Vetter, I., Wilson, D.T., Daly, N.L., Schroeder, C.I., Bourinet, E., Dutertre, S.(2023) FEBS J 290: 3688-3702
- PubMed: 36912793
- DOI: https://doi.org/10.1111/febs.16773
- Primary Citation of Related Structures:
8FEY - PubMed Abstract:
Venom-derived peptides targeting ion channels involved in pain are regarded as a promising alternative to current, and often ineffective, chronic pain treatments. Many peptide toxins are known to specifically and potently block established therapeutic targets, among which the voltage-gated sodium and calcium channels are major contributors. Here, we report on the discovery and characterization of a novel spider toxin isolated from the crude venom of Pterinochilus murinus that shows inhibitory activity at both hNa V 1.7 and hCa V 3.2 channels, two therapeutic targets implicated in pain pathways. Bioassay-guided HPLC fractionation revealed a 36-amino acid peptide with three disulfide bridges named μ/ω-theraphotoxin-Pmu1a (Pmu1a). Following isolation and characterization, the toxin was chemically synthesized and its biological activity was further assessed using electrophysiology, revealing Pmu1a to be a toxin that potently blocks both hNa V 1.7 and hCa V 3. Nuclear magnetic resonance structure determination of Pmu1a shows an inhibitor cystine knot fold that is the characteristic of many spider peptides. Combined, these data show the potential of Pmu1a as a basis for the design of compounds with dual activity at the therapeutically relevant hCa V 3.2 and hNa V 1.7 voltage-gated channels.
Organizational Affiliation:
IBMM, CNRS, ENSCM, Université de Montpellier, France.