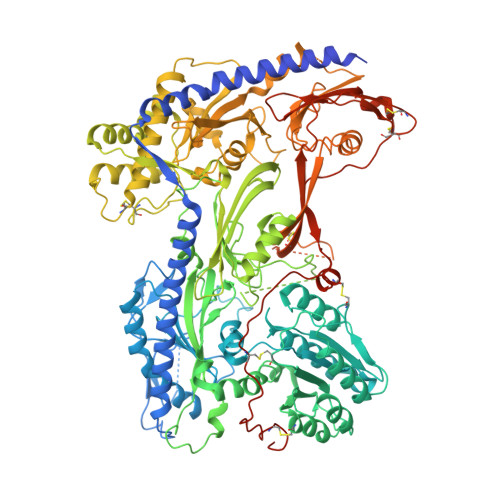
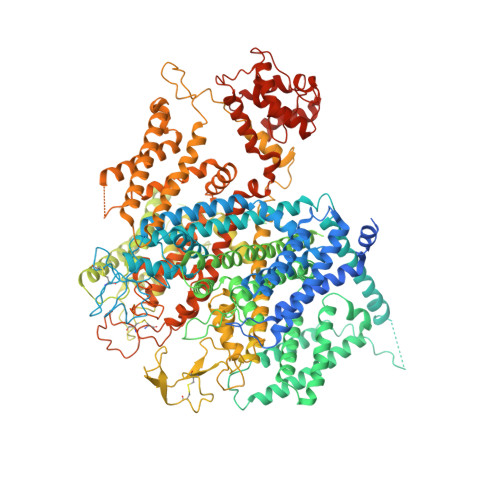
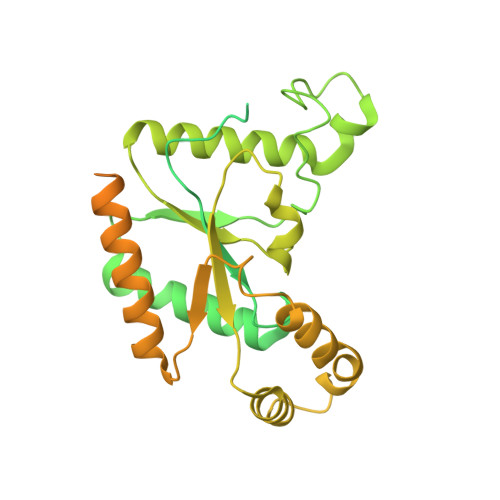
Structural basis for Ca V alpha 2 delta :gabapentin binding.
Chen, Z., Mondal, A., Minor Jr., D.L.(2023) Nat Struct Mol Biol 30: 735-739
- PubMed: 36973510
- DOI: https://doi.org/10.1038/s41594-023-00951-7
- Primary Citation of Related Structures:
8FD7 - PubMed Abstract:
Gabapentinoid drugs for pain and anxiety act on the Ca V α 2 δ-1 and Ca V α 2 δ-2 subunits of high-voltage-activated calcium channels (Ca V 1s and Ca V 2s). Here we present the cryo-EM structure of the gabapentin-bound brain and cardiac Ca V 1.2/Ca V β 3 /Ca V α 2 δ-1 channel. The data reveal a binding pocket in the Ca V α 2 δ-1 dCache1 domain that completely encapsulates gabapentin and define Ca V α 2 δ isoform sequence variations that explain the gabapentin binding selectivity of Ca V α 2 δ-1 and Ca V α 2 δ-2.
- Cardiovascular Research Institute, University of California, San Francisco, CA, USA.
Organizational Affiliation: