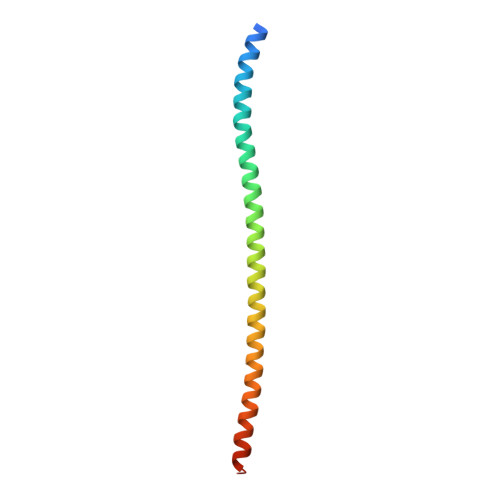Structural insights into plasmalemma vesicle-associated protein (PLVAP): Implications for vascular endothelial diaphragms and fenestrae.
Chang, T.H., Hsieh, F.L., Gu, X., Smallwood, P.M., Kavran, J.M., Gabelli, S.B., Nathans, J.(2023) Proc Natl Acad Sci U S A 120: e2221103120-e2221103120
- PubMed: 36996108
- DOI: https://doi.org/10.1073/pnas.2221103120
- Primary Citation of Related Structures:
8FBY, 8FCF - PubMed Abstract:
In many organs, small openings across capillary endothelial cells (ECs) allow the diffusion of low-molecular weight compounds and small proteins between the blood and tissue spaces. These openings contain a diaphragm composed of radially arranged fibers, and current evidence suggests that a single-span type II transmembrane protein, plasmalemma vesicle-associated protein-1 (PLVAP), constitutes these fibers. Here, we present the three-dimensional crystal structure of an 89-amino acid segment of the PLVAP extracellular domain (ECD) and show that it adopts a parallel dimeric alpha-helical coiled-coil configuration with five interchain disulfide bonds. The structure was solved using single-wavelength anomalous diffraction from sulfur-containing residues (sulfur SAD) to generate phase information. Biochemical and circular dichroism (CD) experiments show that a second PLVAP ECD segment also has a parallel dimeric alpha-helical configuration-presumably a coiled coil-held together with interchain disulfide bonds. Overall, ~2/3 of the ~390 amino acids within the PLVAP ECD adopt a helical configuration, as determined by CD. We also determined the sequence and epitope of MECA-32, an anti-PLVAP antibody. Taken together, these data lend strong support to the model of capillary diaphragms formulated by Tse and Stan in which approximately ten PLVAP dimers are arranged within each 60- to 80-nm-diameter opening like the spokes of a bicycle wheel. Passage of molecules through the wedge-shaped pores is presumably determined both by the length of PLVAP-i.e., the long dimension of the pore-and by the chemical properties of amino acid side chains and N-linked glycans on the solvent-accessible faces of PLVAP.
- Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine, Baltimore, MD 21205.
Organizational Affiliation: