Dog sialic acid esterase (SIAE)
Ide, D., Gorelik, A., Illes, K., Nagar, B.To be published.
Experimental Data Snapshot
Starting Model: other
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sialic acid acetylesterase | 496 | Canis lupus familiaris | Mutation(s): 0 EC: 3.1.1.53 | 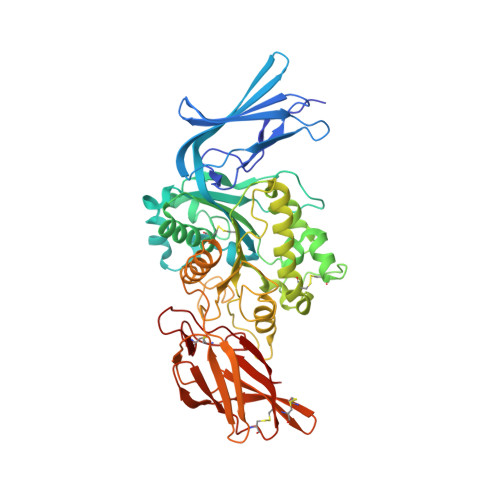 | |
UniProt | |||||
Find proteins for A0A8C0LZX8 (Canis lupus familiaris) Explore A0A8C0LZX8 Go to UniProtKB: A0A8C0LZX8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8C0LZX8 | ||||
Glycosylation | |||||
| Glycosylation Sites: 5 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | B | 6 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G82348BZ GlyCosmos: G82348BZ GlyGen: G82348BZ | |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | C | 4 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G81315DD GlyCosmos: G81315DD GlyGen: G81315DD | |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | F [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 111.702 | α = 90 |
| b = 112.48 | β = 90 |
| c = 122.243 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Canadian Institutes of Health Research (CIHR) | Canada | 250057 |