Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
Harkness, R.W., Ripstein, Z.A., Di Trani, J.M., Kay, L.E.(2023) J Am Chem Soc 145: 13015-13026
- PubMed: 37282495
- DOI: https://doi.org/10.1021/jacs.2c11849
- Primary Citation of Related Structures:
8F0A, 8F0U, 8F1T, 8F1U, 8F21, 8F26 - PubMed Abstract:
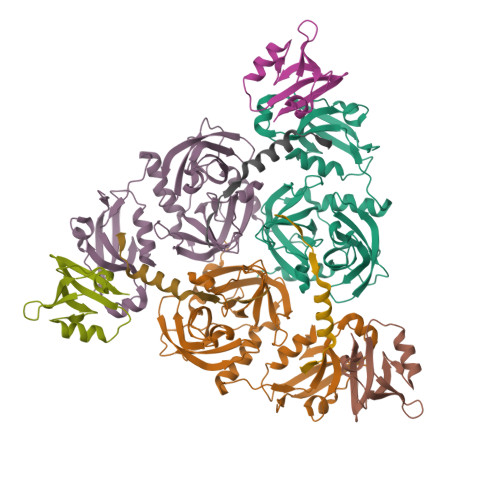
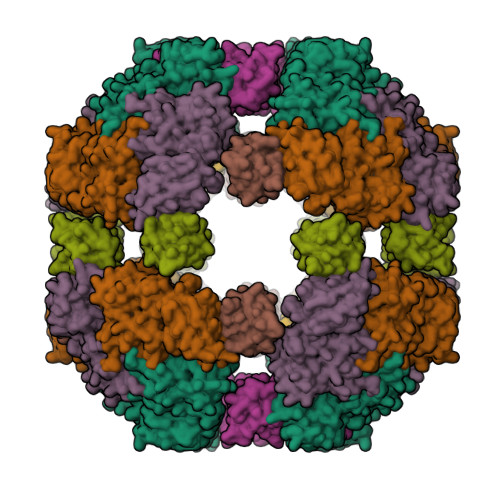
The periplasmic protein DegP, which is implicated in virulence factor transport leading to pathogenicity, is a bi-functional protease and chaperone that helps to maintain protein homeostasis in Gram-negative bacteria and is essential to bacterial survival under stress conditions. To perform these functions, DegP captures clients inside cage-like structures, which we have recently shown to form through the reorganization of high-order preformed apo oligomers, consisting of trimeric building blocks, that are structurally distinct from client-bound cages. Our previous studies suggested that these apo oligomers may allow DegP to encapsulate clients of various sizes under protein folding stresses by forming ensembles that can include extremely large cage particles, but how this occurs remains an open question. To explore the relation between cage and substrate sizes, we engineered a series of DegP clients of increasing hydrodynamic radii and analyzed their influence on DegP cage formation. We used dynamic light scattering and cryogenic electron microscopy to characterize the hydrodynamic properties and structures of the DegP cages that are adopted in response to each client. We present a series of density maps and structural models that include those for novel particles of approximately 30 and 60 monomers. Key interactions between DegP trimers and the bound clients that stabilize the cage assemblies and prime the clients for catalysis are revealed. We also provide evidence that DegP can form cages which approach subcellular organelles in terms of size.
Organizational Affiliation:
Department of Biochemistry, University of Toronto, Toronto M5S 1A8, Canada.