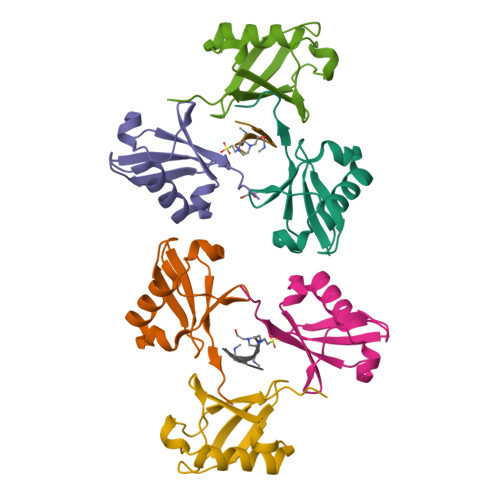
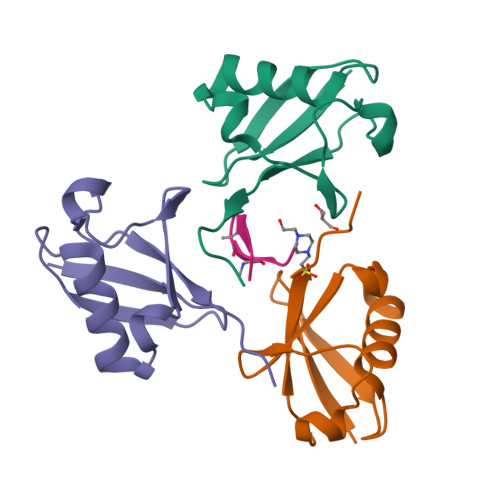
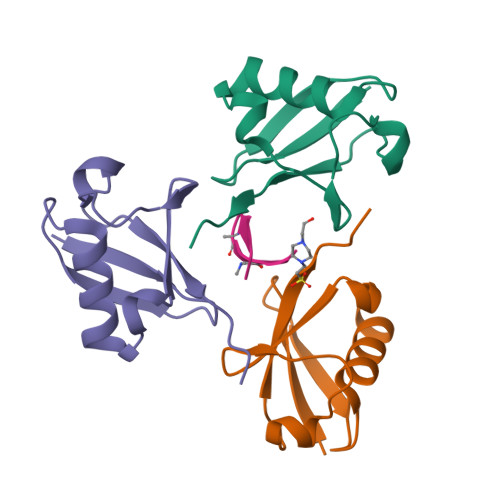
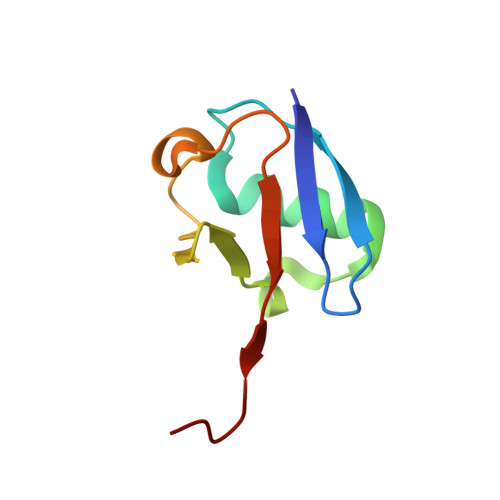
Mechanism of selective recognition of Lys48-linked polyubiquitin by macrocyclic peptide inhibitors of proteasomal degradation.
Lemma, B., Zhang, D., Vamisetti, G.B., Wentz, B.G., Suga, H., Brik, A., Lubkowski, J., Fushman, D.(2023) Nat Commun 14: 7212-7212
- PubMed: 37938554
- DOI: https://doi.org/10.1038/s41467-023-43025-4
- Primary Citation of Related Structures:
8E7O, 8F1F - PubMed Abstract:
Post-translational modification of proteins with polyubiquitin chains is a critical cellular signaling mechanism in eukaryotes with implications in various cellular states and processes. Unregulated ubiquitin-mediated protein degradation can be detrimental to cellular homeostasis, causing numerous diseases including cancers. Recently, macrocyclic peptides were developed that selectively target long Lysine-48-linked polyubiquitin chains (tetra-ubiquitin) to inhibit ubiquitin-proteasome system, leading to attenuation of tumor growth in vivo. However, structural determinants of the chain length and linkage selectivity by these cyclic peptides remained unclear. Here, we uncover the mechanism underlying cyclic peptide's affinity and binding selectivity by combining X-ray crystallography, solution NMR, and biochemical studies. We found that the peptide engages three consecutive ubiquitins that form a ring around the peptide and determined requirements for preferential selection of a specific trimer moiety in longer polyubiquitin chains. The structural insights gained from this work will guide the development of next-generation cyclic peptides with enhanced anti-cancer activity.
Organizational Affiliation:
Center for Biomolecular Structure and Organization, Department of Chemistry and Biochemistry, University of Maryland, College Park, MD, 20742, USA.