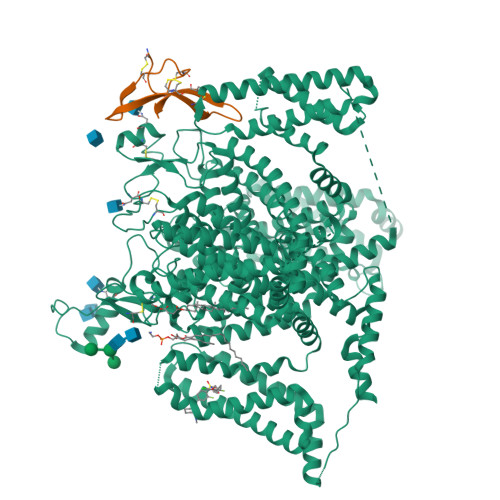
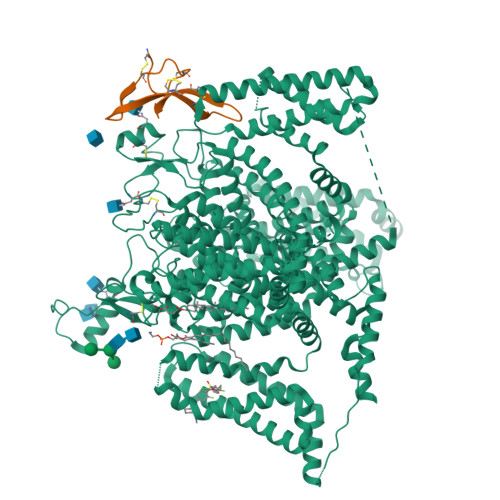
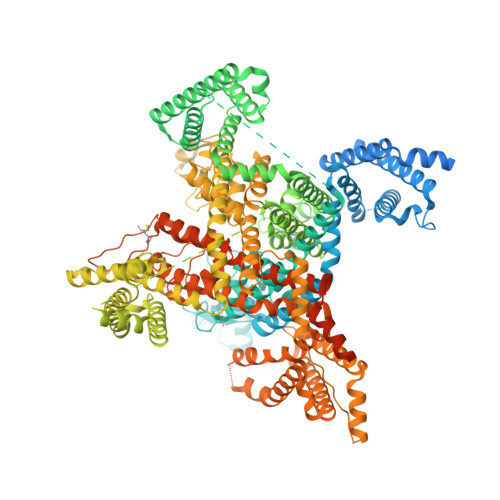
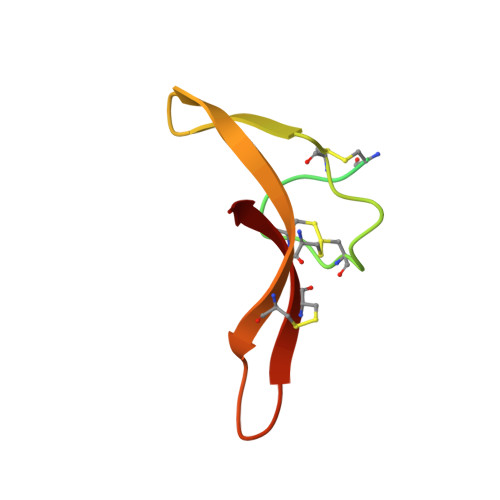
Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Kschonsak, M., Jao, C.C., Arthur, C.P., Rohou, A.L., Bergeron, P., Ortwine, D.F., McKerrall, S.J., Hackos, D.H., Deng, L., Chen, J., Li, T., Dragovich, P.S., Volgraf, M., Wright, M.R., Payandeh, J., Ciferri, C., Tellis, J.C.(2023) Elife 12
- PubMed: 36975198
- DOI: https://doi.org/10.7554/eLife.84151
- Primary Citation of Related Structures:
8F0P, 8F0Q, 8F0R, 8F0S - PubMed Abstract:
The voltage-gated sodium (Na V ) channel Na V 1.7 has been identified as a potential novel analgesic target due to its involvement in human pain syndromes. However, clinically available Na V channel-blocking drugs are not selective among the nine Na V channel subtypes, Na V 1.1-Na V 1.9. Moreover, the two currently known classes of Na V 1.7 subtype-selective inhibitors (aryl- and acylsulfonamides) have undesirable characteristics that may limit their development. To this point understanding of the structure-activity relationships of the acylsulfonamide class of Na V 1.7 inhibitors, exemplified by the clinical development candidate GDC-0310 , has been based solely on a single co-crystal structure of an arylsulfonamide inhibitor bound to voltage-sensing domain 4 (VSD4). To advance inhibitor design targeting the Na V 1.7 channel, we pursued high-resolution ligand-bound Na V 1.7-VSD4 structures using cryogenic electron microscopy (cryo-EM). Here, we report that GDC-0310 engages the Na V 1.7-VSD4 through an unexpected binding mode orthogonal to the arylsulfonamide inhibitor class binding pose, which identifies a previously unknown ligand binding site in Na V channels. This finding enabled the design of a novel hybrid inhibitor series that bridges the aryl- and acylsulfonamide binding pockets and allows for the generation of molecules with substantially differentiated structures and properties. Overall, our study highlights the power of cryo-EM methods to pursue challenging drug targets using iterative and high-resolution structure-guided inhibitor design. This work also underscores an important role of the membrane bilayer in the optimization of selective Na V channel modulators targeting VSD4.
Organizational Affiliation:
Genentech Inc, Structural Biology, South San Francisco, United States.