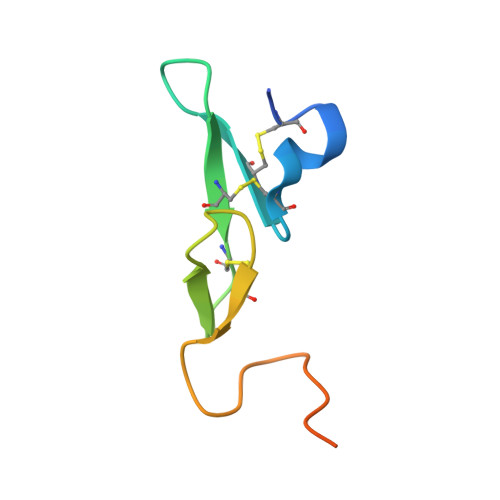
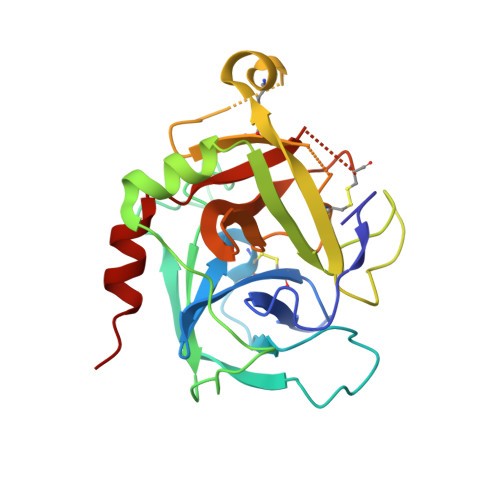
An RNA aptamer exploits exosite-dependent allostery to achieve specific inhibition of coagulation factor IXa.
Kolyadko, V.N., Layzer, J.M., Perry, K., Sullenger, B.A., Krishnaswamy, S.(2024) Proc Natl Acad Sci U S A 121: e2401136121-e2401136121
- PubMed: 38985762
- DOI: https://doi.org/10.1073/pnas.2401136121
- Primary Citation of Related Structures:
8EPC, 8EPH, 8EPK - PubMed Abstract:
Hemostasis relies on a reaction network of serine proteases and their cofactors to form a blood clot. Coagulation factor IXa (protease) plays an essential role in hemostasis as evident from the bleeding disease associated with its absence. RNA aptamers specifically targeting individual coagulation factors have potential as anticoagulants and as probes of the relationship between structure and function. Here, we report X-ray structures of human factor IXa without a ligand bound to the active site either in the apo-form or in complex with an inhibitory aptamer specific for factor IXa. The aptamer binds to an exosite in the catalytic domain and allosterically distorts the active site. Our studies reveal a conformational ensemble of IXa states, wherein large movements of Trp 215 near the active site drive functional transitions between the closed (aptamer-bound), latent (apo), and open (substrate-bound) states. The latent state of the apo-enzyme may bear on the uniquely poor catalytic activity of IXa compared to other coagulation proteases. The exosite, to which the aptamer binds, has been implicated in binding VIIIa and heparin, both of which regulate IXa function. Our findings reveal the importance of exosite-driven allosteric modulation of IXa function and new strategies to rebalance hemostasis for therapeutic gain.
Organizational Affiliation:
Division of Hematology, Department of Pediatrics, Children's Hospital of Philadelphia, Philadelphia, PA 19104.