Allosteric mechanism of transcription inhibition by NusG-dependent pausing of RNA polymerase.
Vishwakarma, R.K., Qayyum, M.Z., Babitzke, P., Murakami, K.S.(2023) Proc Natl Acad Sci U S A 120: e2218516120-e2218516120
- PubMed: 36745813
- DOI: https://doi.org/10.1073/pnas.2218516120
- Primary Citation of Related Structures:
8EHQ, 8EJ3, 8EOE, 8EOF, 8EOS, 8EOT, 8EXY - PubMed Abstract:
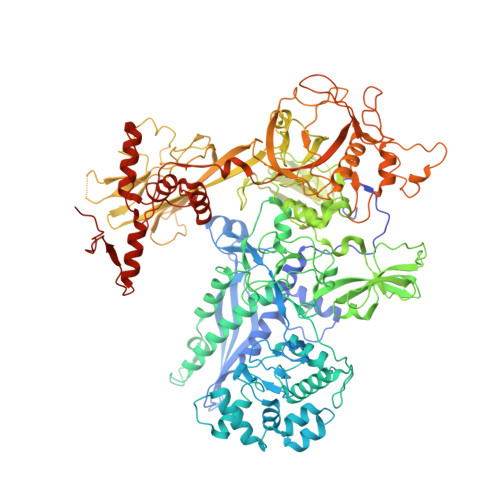
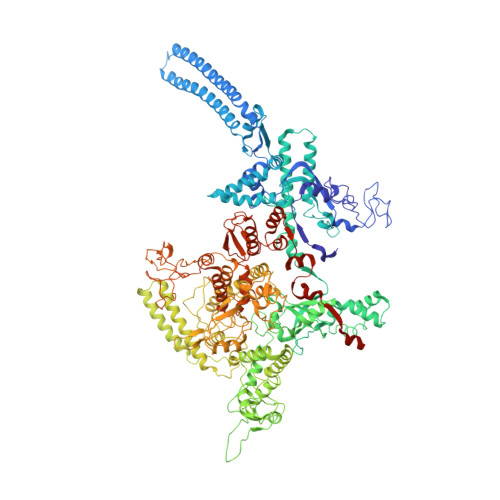
NusG is a transcription elongation factor that stimulates transcription pausing in Gram+ bacteria including B. subtilis by sequence-specific interaction with a conserved pause-inducing - 11 TTNTTT -6 motif found in the non-template DNA (ntDNA) strand within the transcription bubble. To reveal the structural basis of NusG-dependent pausing, we determined a cryo-EM structure of a paused transcription complex (PTC) containing RNA polymerase (RNAP), NusG, and the TTNTTT motif in the ntDNA strand. The interaction of NusG with the ntDNA strand rearranges the transcription bubble by positioning three consecutive T residues in a cleft between NusG and the β-lobe domain of RNAP. We revealed that the RNAP swivel module rotation (swiveling), which widens (swiveled state) and narrows (non-swiveled state) a cleft between NusG and the β-lobe, is an intrinsic motion of RNAP and is directly linked to trigger loop (TL) folding, an essential conformational change of all cellular RNAPs for the RNA synthesis reaction. We also determined cryo-EM structures of RNAP escaping from the paused transcription state. These structures revealed the NusG-dependent pausing mechanism by which NusG-ntDNA interaction inhibits the transition from swiveled to non-swiveled states, thereby preventing TL folding and RNA synthesis allosterically. This motion is also reduced by the formation of an RNA hairpin within the RNA exit channel. Thus, the pause half-life can be modulated by the strength of the NusG-ntDNA interaction and/or the stability of the RNA hairpin. NusG residues that interact with the TTNTTT motif are widely conserved in bacteria, suggesting that NusG-dependent pausing is widespread.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Center for RNA Molecular Biology, Pennsylvania State University, University Park, PA 16802.