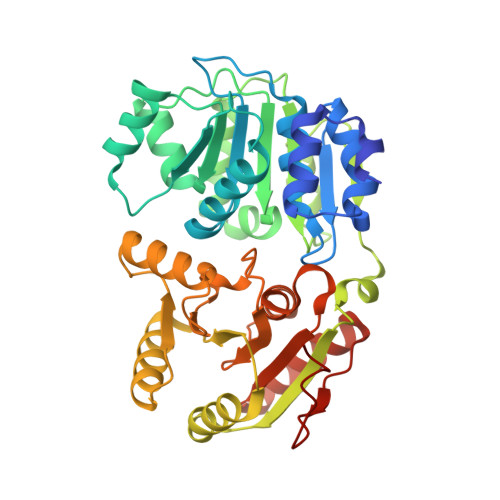
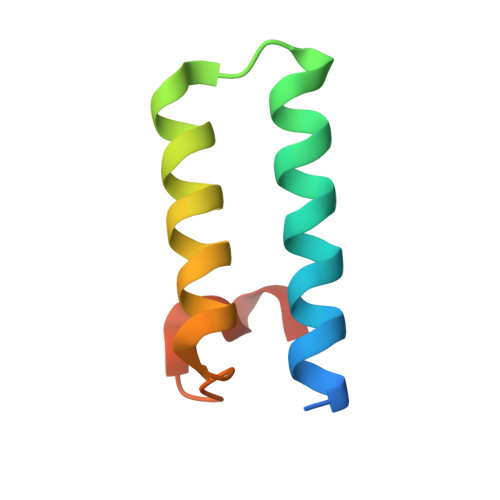
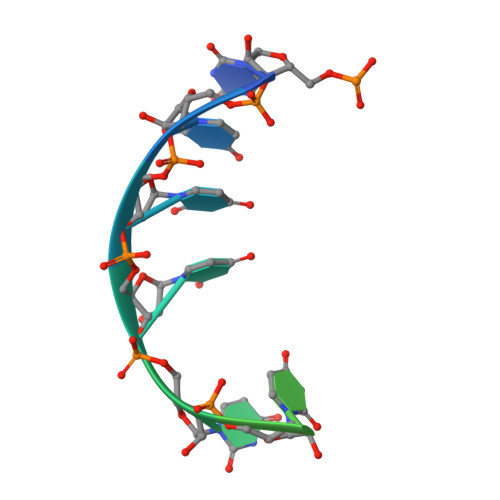
Structural basis for high-order complex of SARNP and DDX39B to facilitate mRNP assembly.
Xie, Y., Gao, S., Zhang, K., Bhat, P., Clarke, B.P., Batten, K., Mei, M., Gazzara, M., Shay, J.W., Lynch, K.W., Angelos, A.E., Hill, P.S., Ivey, A.L., Fontoura, B., Ren, Y.(2023) Cell Rep 42: 112988-112988
- PubMed: 37578863
- DOI: https://doi.org/10.1016/j.celrep.2023.112988
- Primary Citation of Related Structures:
8ENK - PubMed Abstract:
mRNA in eukaryotic cells is packaged into highly compacted ribonucleoprotein particles (mRNPs) in the nucleus and exported to the cytoplasm for translation. mRNP packaging and export require the evolutionarily conserved transcription-export (TREX) complex. TREX facilitates loading of various RNA-binding proteins on mRNA through the action of its DDX39B subunit. SARNP (Tho1 [transcriptional defect of Hpr1 by overexpression 1] in yeast) is shown to interact with DDX39B and affect mRNA export. The molecular mechanism of how SARNP recognizes DDX39B and functions in mRNP assembly is unclear. Here, we determine the crystal structure of a Tho1/DDX39B/RNA complex, revealing a multivalent interaction mediated by tandem DDX39B interacting motifs in SARNP/Tho1. The high-order complex of SARNP and DDX39B is evolutionarily conserved, and human SARNP can engage with five DDX39B molecules. RNA sequencing (RNA-seq) from SARNP knockdown cells shows the most affected RNAs in export are GC rich. Our work suggests the role of the high-order SARNP/DDX39B/RNA complex in mRNP assembly and export.
Organizational Affiliation:
Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, TN 37232-0146, USA; Center for Structural Biology, Vanderbilt University School of Medicine, Nashville, TN 37232-0146, USA.