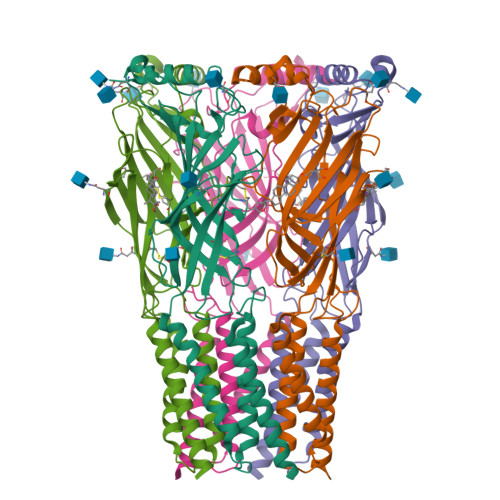
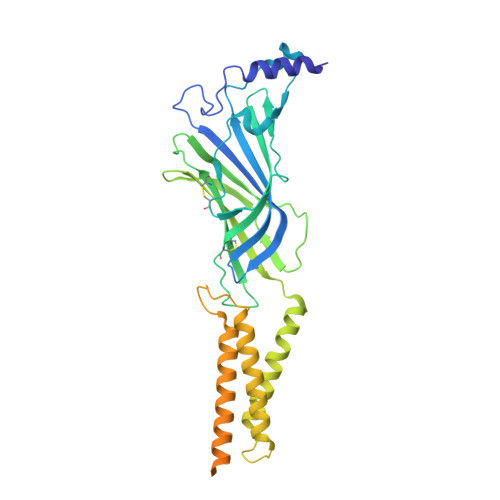
Sensory specializations drive octopus and squid behaviour.
Kang, G., Allard, C.A.H., Valencia-Montoya, W.A., van Giesen, L., Kim, J.J., Kilian, P.B., Bai, X., Bellono, N.W., Hibbs, R.E.(2023) Nature 616: 378-383
- PubMed: 37045917
- DOI: https://doi.org/10.1038/s41586-023-05808-z
- Primary Citation of Related Structures:
8EIS, 8EIZ - PubMed Abstract:
The evolution of new traits enables expansion into new ecological and behavioural niches. Nonetheless, demonstrated connections between divergence in protein structure, function and lineage-specific behaviours remain rare. Here we show that both octopus and squid use cephalopod-specific chemotactile receptors (CRs) to sense their respective marine environments, but structural adaptations in these receptors support the sensation of specific molecules suited to distinct physiological roles. We find that squid express ancient CRs that more closely resemble related nicotinic acetylcholine receptors, whereas octopuses exhibit a more recent expansion in CRs consistent with their elaborated 'taste by touch' sensory system. Using a combination of genetic profiling, physiology and behavioural analyses, we identify the founding member of squid CRs that detects soluble bitter molecules that are relevant in ambush predation. We present the cryo-electron microscopy structure of a squid CR and compare this with octopus CRs 1 and nicotinic receptors 2 . These analyses demonstrate an evolutionary transition from an ancestral aromatic 'cage' that coordinates soluble neurotransmitters or tastants to a more recent octopus CR hydrophobic binding pocket that traps insoluble molecules to mediate contact-dependent chemosensation. Thus, our study provides a foundation for understanding how adaptation of protein structure drives the diversification of organismal traits and behaviour.
Organizational Affiliation:
Department of Neuroscience, University of Texas Southwestern Medical Center, Dallas, TX, USA.