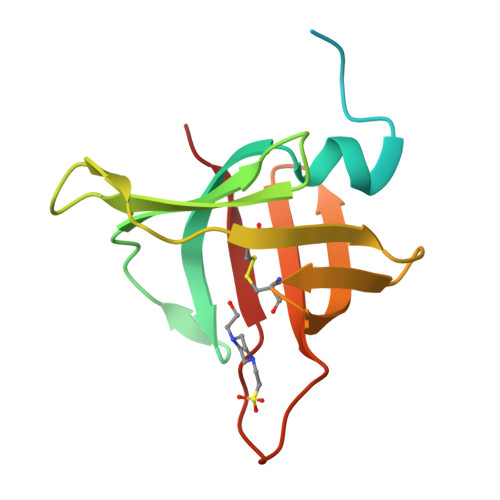
A unique network of attack, defence and competence on the outer membrane of the periodontitis pathogen Tannerella forsythia.
Ksiazek, M., Goulas, T., Mizgalska, D., Rodriguez-Banqueri, A., Eckhard, U., Veillard, F., Waligorska, I., Benedyk-Machaczka, M., Sochaj-Gregorczyk, A.M., Madej, M., Thogersen, I.B., Enghild, J.J., Cuppari, A., Arolas, J.L., de Diego, I., Lopez-Pelegrin, M., Garcia-Ferrer, I., Guevara, T., Dive, V., Zani, M.L., Moreau, T., Potempa, J., Gomis-Ruth, F.X.(2023) Chem Sci 14: 869-888
- PubMed: 36755705
- DOI: https://doi.org/10.1039/d2sc04166a
- Primary Citation of Related Structures:
8B2M, 8B2N, 8B2Q, 8EHB, 8EHC, 8EHD, 8EHE - PubMed Abstract:
Periodontopathogenic Tannerella forsythia uniquely secretes six peptidases of disparate catalytic classes and families that operate as virulence factors during infection of the gums, the KLIKK-peptidases. Their coding genes are immediately downstream of novel ORFs encoding the 98-132 residue potempins (Pot) A, B1, B2, C, D and E. These are outer-membrane-anchored lipoproteins that specifically and potently inhibit the respective downstream peptidase through stable complexes that protect the outer membrane of T. forsythia , as shown in vivo . Remarkably, PotA also contributes to bacterial fitness in vivo and specifically inhibits matrix metallopeptidase (MMP) 12, a major defence component of oral macrophages, thus featuring a novel and highly-specific physiological MMP inhibitor. Information from 11 structures and high-confidence homology models showed that the potempins are distinct β-barrels with either a five-stranded OB-fold (PotA, PotC and PotD) or an eight-stranded up-and-down fold (PotE, PotB1 and PotB2), which are novel for peptidase inhibitors. Particular loops insert like wedges into the active-site cleft of the genetically-linked peptidases to specifically block them either via a new "bilobal" or the classic "standard" mechanism of inhibition. These results discover a unique, tightly-regulated proteolytic armamentarium for virulence and competence, the KLIKK-peptidase/potempin system.
Organizational Affiliation:
Department of Microbiology, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University Gronostajowa 7 Kraków 30-387 Poland jan.potempa@icloud.com.