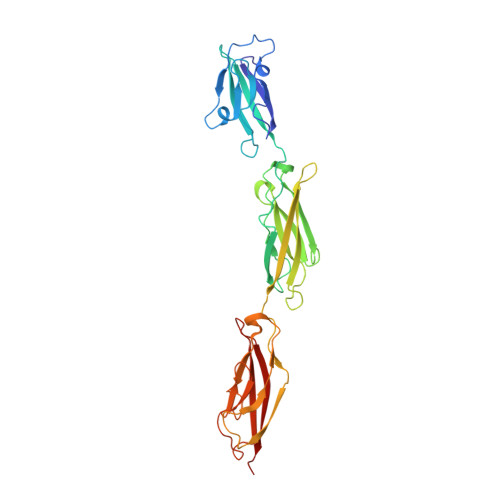
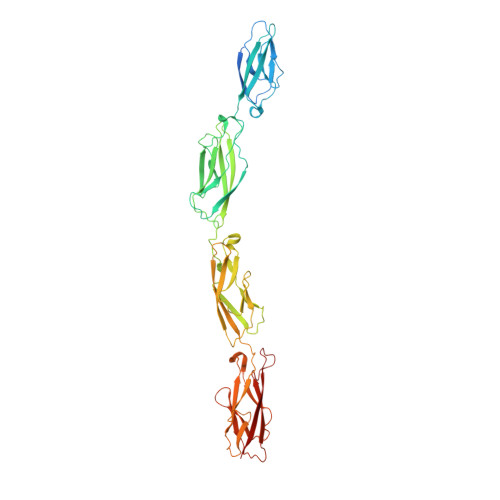Structure of the planar cell polarity cadherins Fat4 and Dachsous1.
Medina, E., Easa, Y., Lester, D.K., Lau, E.K., Sprinzak, D., Luca, V.C.(2023) Nat Commun 14: 891-891
- PubMed: 36797229
- DOI: https://doi.org/10.1038/s41467-023-36435-x
- Primary Citation of Related Structures:
8EGW, 8EGX - PubMed Abstract:
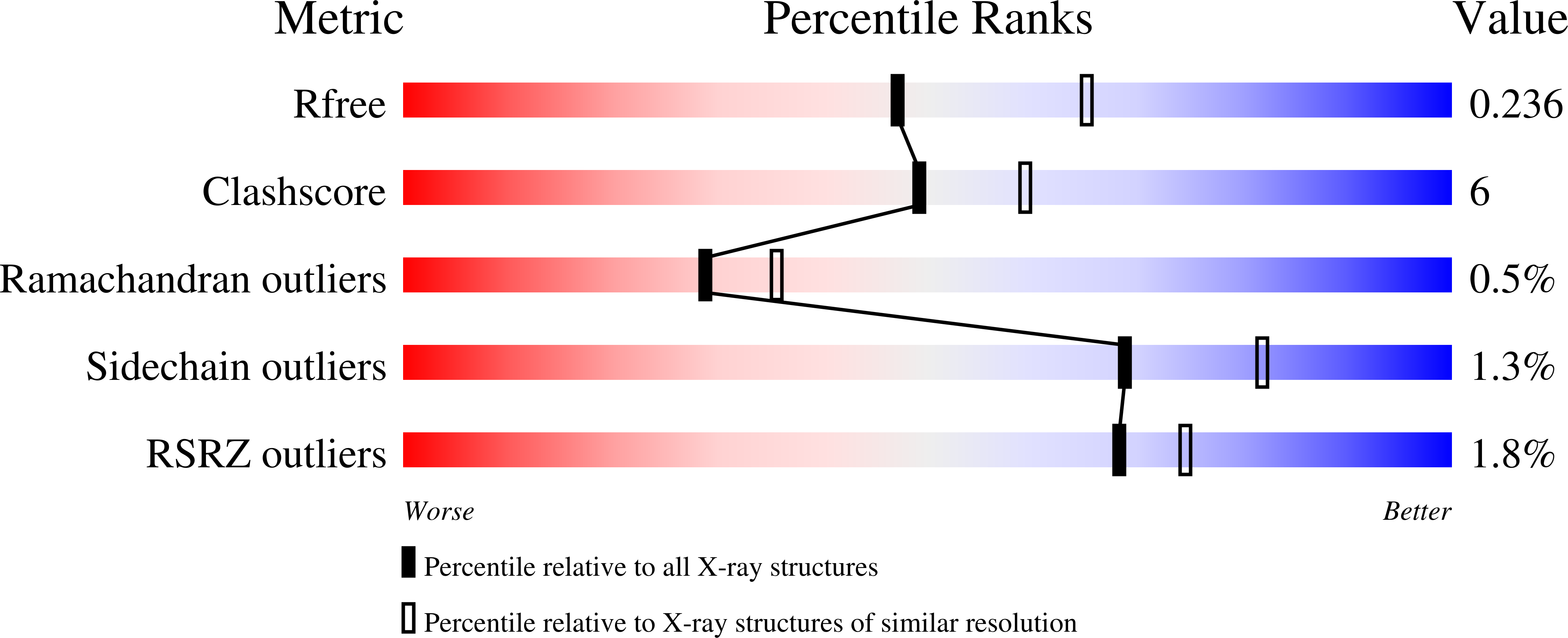
The atypical cadherins Fat and Dachsous are key regulators of cell growth and animal development. In contrast to classical cadherins, which form homophilic interactions to segregate cells, Fat and Dachsous cadherins form heterophilic interactions to induce cell polarity within tissues. Here, we determine the co-crystal structure of the human homologs Fat4 and Dachsous1 (Dchs1) to establish the molecular basis for Fat-Dachsous interactions. The binding domains of Fat4 and Dchs1 form an extended interface along extracellular cadherin (EC) domains 1-4 of each protein. Biophysical measurements indicate that Fat4-Dchs1 affinity is among the highest reported for cadherin superfamily members, which is attributed to an extensive network of salt bridges not present in structurally similar protocadherin homodimers. Furthermore, modeling suggests that unusual extracellular phosphorylation modifications directly modulate Fat-Dachsous binding by introducing charged contacts across the interface. Collectively, our analyses reveal how the molecular architecture of Fat4-Dchs1 enables them to form long-range, high-affinity interactions to maintain planar cell polarity.
Organizational Affiliation:
Department of Drug Discovery, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, 33602, USA.