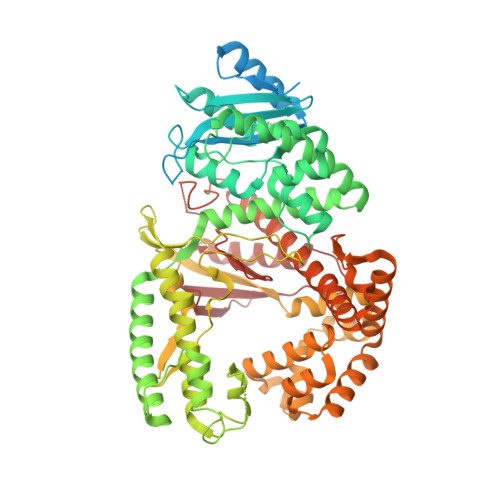
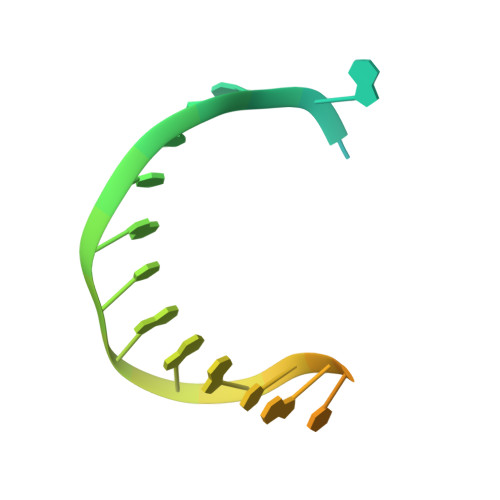
Structural basis of DNA polymerase theta mediated DNA end joining.
Li, C., Zhu, H., Jin, S., Maksoud, L.M., Jain, N., Sun, J., Gao, Y.(2023) Nucleic Acids Res 51: 463-474
- PubMed: 36583344
- DOI: https://doi.org/10.1093/nar/gkac1201
- Primary Citation of Related Structures:
8EF9, 8EFC, 8EFK - PubMed Abstract:
DNA polymerase θ (Pol θ) plays an essential role in the microhomology-mediated end joining (MMEJ) pathway for repairing DNA double-strand breaks. However, the mechanisms by which Pol θ recognizes microhomologous DNA ends and performs low-fidelity DNA synthesis remain unclear. Here, we present cryo-electron microscope structures of the polymerase domain of Lates calcarifer Pol θ with long and short duplex DNA at up to 2.4 Å resolution. Interestingly, Pol θ binds to long and short DNA substrates similarly, with extensive interactions around the active site. Moreover, Pol θ shares a similar active site as high-fidelity A-family polymerases with its finger domain well-closed but differs in having hydrophilic residues surrounding the nascent base pair. Computational simulations and mutagenesis studies suggest that the unique insertion loops of Pol θ help to stabilize short DNA binding and assemble the active site for MMEJ repair. Taken together, our results illustrate the structural basis of Pol θ-mediated MMEJ.
Organizational Affiliation:
Department of Biosciences, Rice University, Houston, TX 77005, USA.