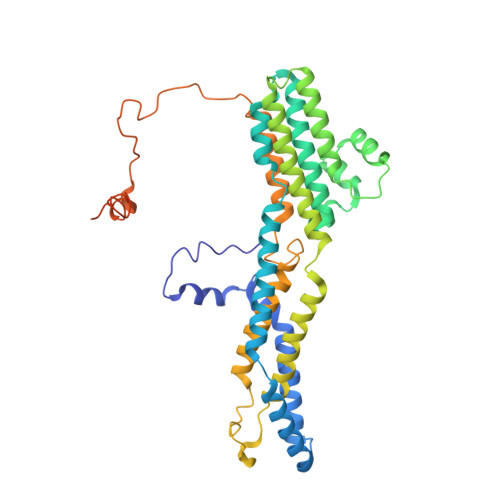
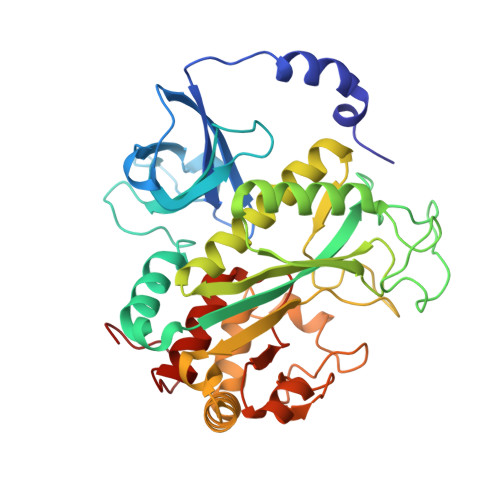
Bestrophin-2 and glutamine synthetase form a complex for glutamate release.
Owji, A.P., Yu, K., Kittredge, A., Wang, J., Zhang, Y., Yang, T.(2022) Nature 611: 180-187
- PubMed: 36289327
- DOI: https://doi.org/10.1038/s41586-022-05373-x
- Primary Citation of Related Structures:
8ECY - PubMed Abstract:
Bestrophin-2 (BEST2) is a member of the bestrophin family of calcium-activated anion channels that has a critical role in ocular physiology 1-4 . Here we uncover a directional permeability of BEST2 to glutamate that heavily favours glutamate exit, identify glutamine synthetase (GS) as a binding partner of BEST2 in the ciliary body of the eye, and solve the structure of the BEST2-GS complex. BEST2 reduces cytosolic GS activity by tethering GS to the cell membrane. GS extends the ion conducting pathway of BEST2 through its central cavity and inhibits BEST2 channel function in the absence of intracellular glutamate, but sensitizes BEST2 to intracellular glutamate, which promotes the opening of BEST2 and thus relieves the inhibitory effect of GS. We demonstrate the physiological role of BEST2 in conducting chloride and glutamate and the influence of GS in non-pigmented ciliary epithelial cells. Together, our results reveal a novel mechanism of glutamate release through BEST2-GS.
Organizational Affiliation:
Department of Ophthalmology, Columbia University, New York, NY, USA.