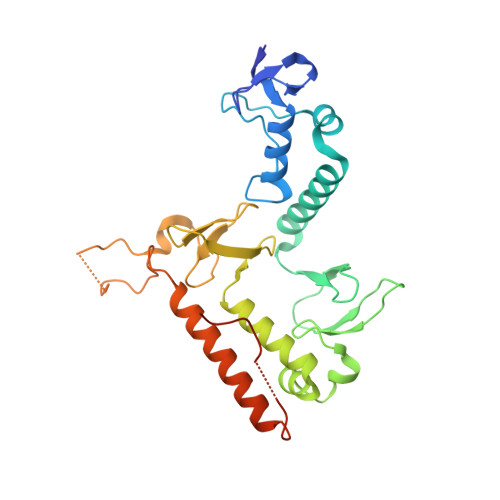
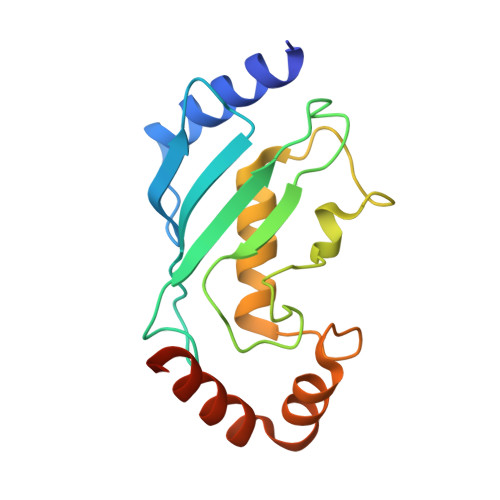
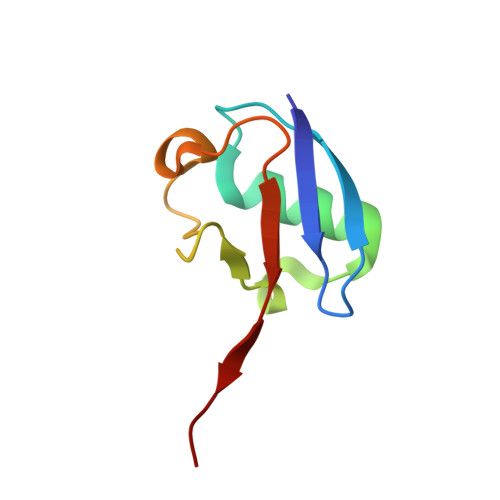
The unifying catalytic mechanism of the RING-between-RING E3 ubiquitin ligase family.
Wang, X.S., Cotton, T.R., Trevelyan, S.J., Richardson, L.W., Lee, W.T., Silke, J., Lechtenberg, B.C.(2023) Nat Commun 14: 168-168
- PubMed: 36631489
- DOI: https://doi.org/10.1038/s41467-023-35871-z
- Primary Citation of Related Structures:
8EAZ, 8EB0 - PubMed Abstract:
The RING-between-RING (RBR) E3 ubiquitin ligase family in humans comprises 14 members and is defined by a two-step catalytic mechanism in which ubiquitin is first transferred from an E2 ubiquitin-conjugating enzyme to the RBR active site and then to the substrate. To define the core features of this catalytic mechanism, we here structurally and biochemically characterise the two RBRs HOIL-1 and RNF216. Crystal structures of both enzymes in their RBR/E2-Ub/Ub transthiolation complexes capturing the first catalytic step, together with complementary functional experiments, reveal the defining features of the RBR catalytic mechanism. RBRs catalyse ubiquitination via a conserved transthiolation complex structure that enables efficient E2-to-RBR ubiquitin transfer. Our data also highlight a conserved RBR allosteric activation mechanism by distinct ubiquitin linkages that suggests RBRs employ a feed-forward mechanism. We finally identify that the HOIL-1 RING2 domain contains an unusual Zn2/Cys6 binuclear cluster that is required for catalytic activity and substrate ubiquitination.
Organizational Affiliation:
Ubiquitin Signalling Division, The Walter and Eliza Hall Institute of Medical Research, Parkville, Victoria, 3052, Australia.