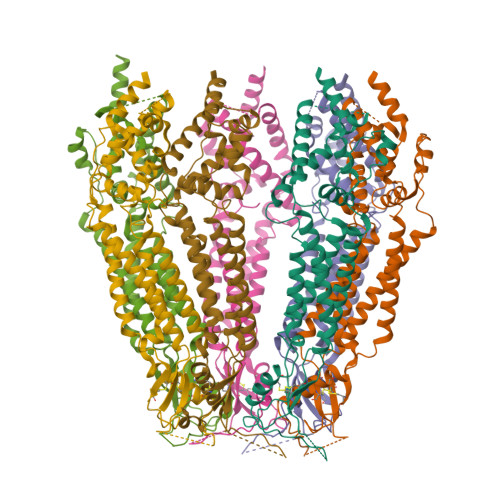
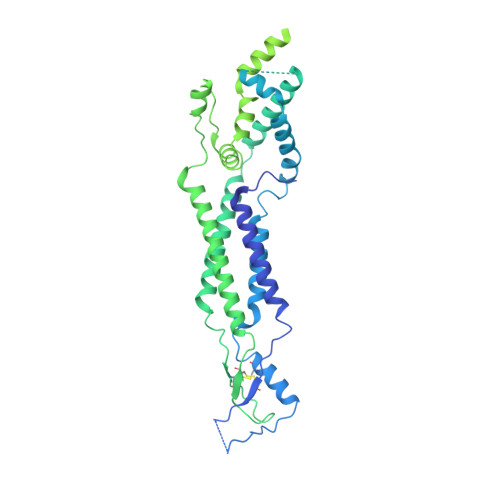
Cryo-EM structures of a LRRC8 chimera with native functional properties reveal heptameric assembly.
Takahashi, H., Yamada, T., Denton, J.S., Strange, K., Karakas, E.(2023) Elife 12
- PubMed: 36897307
- DOI: https://doi.org/10.7554/eLife.82431
- Primary Citation of Related Structures:
8DXN, 8DXO, 8DXP, 8DXQ, 8DXR - PubMed Abstract:
Volume-regulated anion channels (VRACs) mediate volume regulatory Cl - and organic solute efflux from vertebrate cells. VRACs are heteromeric assemblies of LRRC8A-E proteins with unknown stoichiometries. Homomeric LRRC8A and LRRC8D channels have a small pore, hexameric structure. However, these channels are either non-functional or exhibit abnormal regulation and pharmacology, limiting their utility for structure-function analyses. We circumvented these limitations by developing novel homomeric LRRC8 chimeric channels with functional properties consistent with those of native VRAC/LRRC8 channels. We demonstrate here that the LRRC8C-LRRC8A(IL1 25 ) chimera comprising LRRC8C and 25 amino acids unique to the first intracellular loop (IL1) of LRRC8A has a heptameric structure like that of homologous pannexin channels. Unlike homomeric LRRC8A and LRRC8D channels, heptameric LRRC8C-LRRC8A(IL1 25 ) channels have a large-diameter pore similar to that estimated for native VRACs, exhibit normal DCPIB pharmacology, and have higher permeability to large organic anions. Lipid-like densities are located between LRRC8C-LRRC8A(IL1 25 ) subunits and occlude the channel pore. Our findings provide new insights into VRAC/LRRC8 channel structure and suggest that lipids may play important roles in channel gating and regulation.
Organizational Affiliation:
Department of Molecular Physiology and Biophysics, School of Medicine, Vanderbilt University, Nashville, United States.