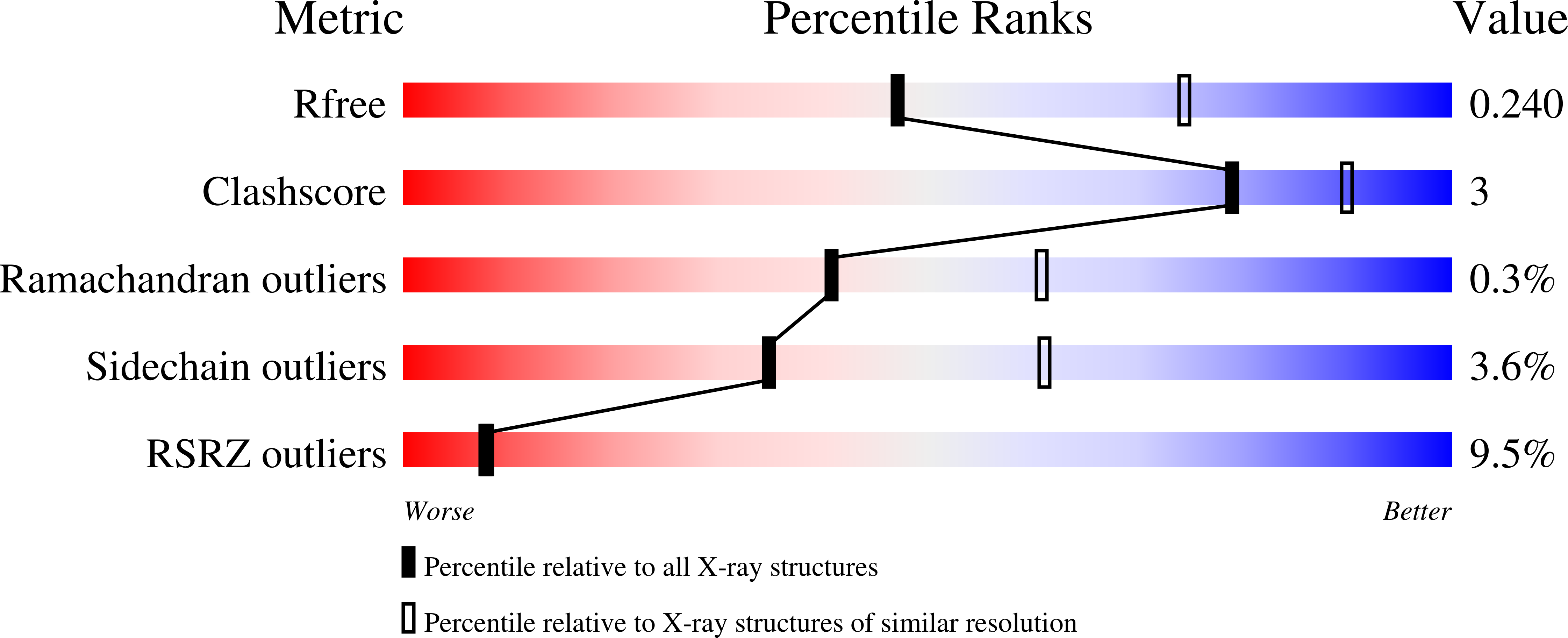
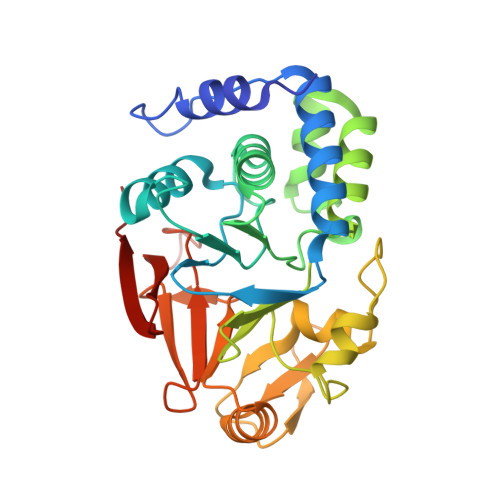
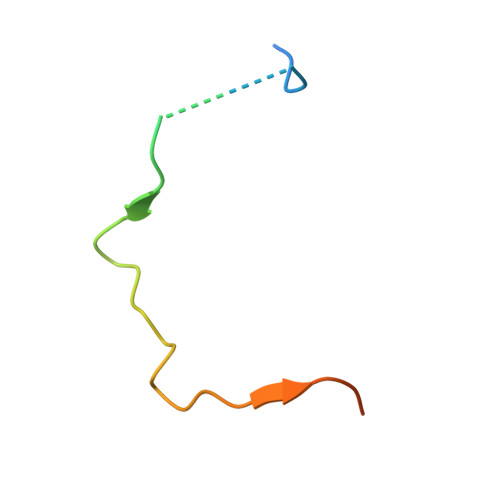
Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Srivastava, G., Choy, M.S., Bolik-Coulon, N., Page, R., Peti, W.(2023) Nat Commun 14: 1798-1798
- PubMed: 37002212
- DOI: https://doi.org/10.1038/s41467-023-37372-5
- Primary Citation of Related Structures:
8DWK, 8DWL - PubMed Abstract:
To achieve substrate specificity, protein phosphate 1 (PP1) forms holoenzymes with hundreds of regulatory and inhibitory proteins. Inhibitor-3 (I3) is an ancient inhibitor of PP1 with putative roles in PP1 maturation and the regulation of PP1 activity. Here, we show that I3 residues 27-68 are necessary and sufficient for PP1 binding and inhibition. In addition to a canonical RVxF motif, which is shared by nearly all PP1 regulators and inhibitors, and a non-canonical SILK motif, I3 also binds PP1 via multiple basic residues that bind directly in the PP1 acidic substrate binding groove, an interaction that provides a blueprint for how substrates bind this groove for dephosphorylation. Unexpectedly, this interaction positions a CCC (cys-cys-cys) motif to bind directly across the PP1 active site. Using biophysical and inhibition assays, we show that the I3 CCC motif binds and inhibits PP1 in an unexpected dynamic, fuzzy manner, via transient engagement of the PP1 active site metals. Together, these data not only provide fundamental insights into the mechanisms by which IDP protein regulators of PP1 achieve inhibition, but also shows that fuzzy interactions between IDPs and their folded binding partners, in addition to enhancing binding affinity, can also directly regulate enzyme activity.
Organizational Affiliation:
Department of Molecular Biology and Biophysics, University of Connecticut Health Center, Farmington, CT, USA.