The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Wang, W., Pyle, A.M.(2022) Mol Cell 82: 4131
- PubMed: 36272408
- DOI: https://doi.org/10.1016/j.molcel.2022.09.029
- Primary Citation of Related Structures:
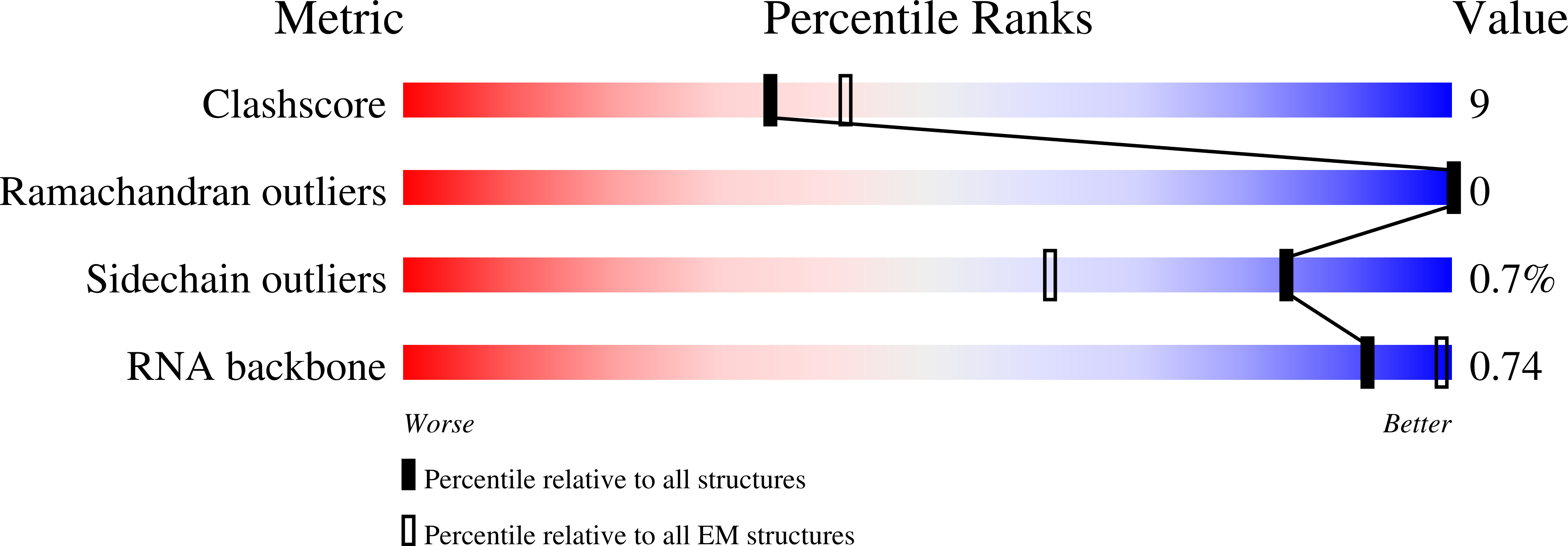
7TNX, 7TNY, 7TNZ, 7TO0, 7TO1, 7TO2, 8DVR, 8DVS, 8DVU - PubMed Abstract:
RIG-I is an essential innate immune receptor for detecting and responding to infection by RNA viruses. RIG-I specifically recognizes the unique molecular features of viral RNA molecules and selectively distinguishes them from closely related RNAs abundant in host cells. The physical basis for this exquisite selectivity is revealed through a series of high-resolution cryo-EM structures of RIG-I in complex with host and viral RNA ligands. These studies demonstrate that RIG-I actively samples double-stranded RNAs in the cytoplasm and distinguishes them by adopting two different types of protein folds. Upon binding viral RNA, RIG-I adopts a high-affinity conformation that is conducive to signaling, while host RNA induces an autoinhibited conformation that stimulates RNA release. By coupling protein folding with RNA binding selectivity, RIG-I distinguishes RNA molecules that differ by as little as one phosphate group, thereby explaining the molecular basis for selective antiviral sensing and the induction of autoimmunity upon RIG-I dysregulation.
Organizational Affiliation:
Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06511, USA; Howard Hughes Medical Institute, Yale University, New Haven, CT 06520, USA.