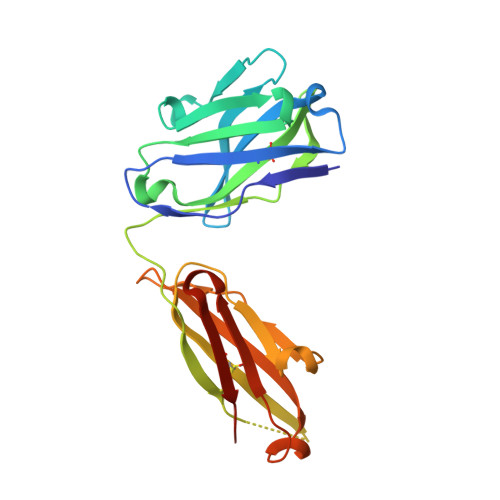
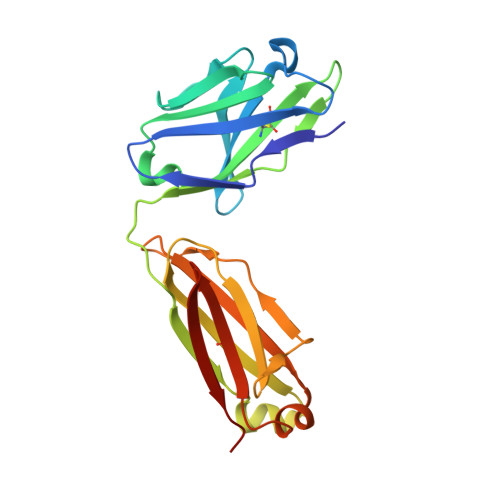
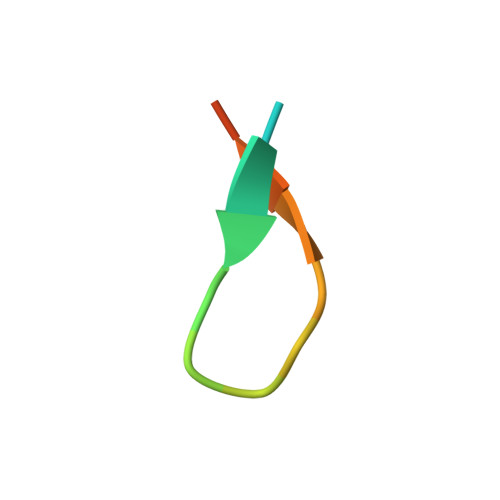
Gonococcal Mimitope Vaccine Candidate Forms a Beta-Hairpin Turn and Binds Hydrophobically to a Therapeutic Monoclonal Antibody.
Beernink, P.T., Di Carluccio, C., Marchetti, R., Cerofolini, L., Carillo, S., Cangiano, A., Cowieson, N., Bones, J., Molinaro, A., Paduano, L., Fragai, M., Beernink, B.P., Gulati, S., Shaughnessy, J., Rice, P.A., Ram, S., Silipo, A.(2024) JACS Au 4: 2617-2629
- PubMed: 39055159
- DOI: https://doi.org/10.1021/jacsau.4c00359
- Primary Citation of Related Structures:
8DOZ, 8DUZ - PubMed Abstract:
The spread of multidrug-resistant strains of Neisseria gonorrhoeae , the etiologic agent of gonorrhea, represents a global health emergency. Therefore, the development of a safe and effective vaccine against gonorrhea is urgently needed. In previous studies, murine monoclonal antibody (mAb) 2C7 was raised against gonococcal lipooligosaccharide (LOS). mAb 2C7 elicits complement-dependent bactericidal activity against gonococci, and its glycan epitope is expressed by almost every clinical isolate. Furthermore, we identified a peptide, cyclic peptide 2 (CP2) that mimicked the 2C7 LOS epitope, elicited bactericidal antibodies in mice, and actively protected in a mouse vaginal colonization model. In this study, we performed structural analyses of mAb 2C7 and its complex with the CP2 peptide by X-ray crystallography, NMR spectroscopy, and molecular dynamics (MD) simulations. The crystal structure of Fab 2C7 bound to CP2 showed that the peptide adopted a beta-hairpin conformation and bound the Fab primarily through hydrophobic interactions. We employed NMR spectroscopy and MD simulations to map the 2C7 epitope and identify the bioactive conformation of CP2. We also used small-angle X-ray scattering (SAXS) and native mass spectrometry to obtain further information about the shape and assembly state of the complex. Collectively, our new structural information suggests strategies for humanizing mAb 2C7 as a therapeutic against gonococcal infection and for optimizing peptide CP2 as a vaccine antigen.
Organizational Affiliation:
Department of Pediatrics, University of California San Francisco, 5700 Martin Luther King Jr. Way, Oakland, California 94609, United States.