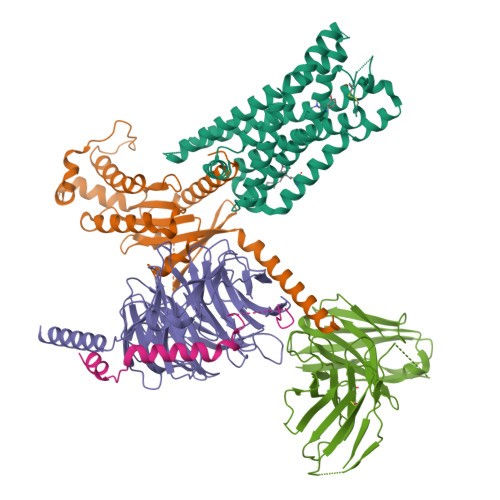
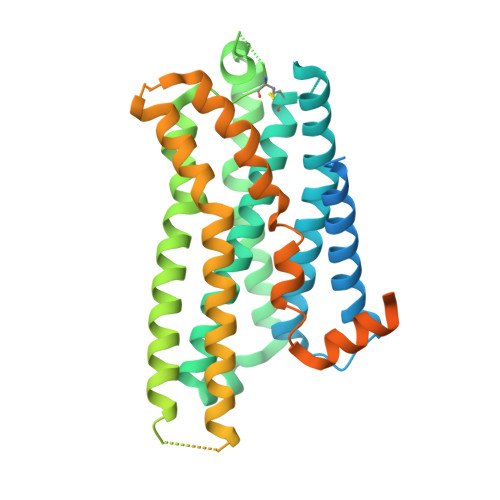
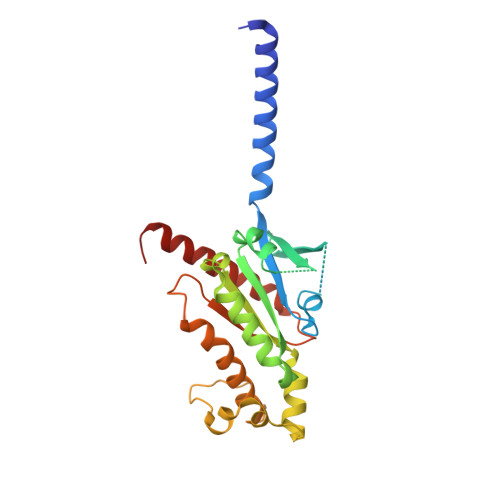
Molecular insights into the regulation of constitutive activity by RNA editing of 5HT 2C serotonin receptors.
Gumpper, R.H., Fay, J.F., Roth, B.L.(2022) Cell Rep 40: 111211-111211
- PubMed: 35977511
- DOI: https://doi.org/10.1016/j.celrep.2022.111211
- Primary Citation of Related Structures:
8DPF, 8DPG, 8DPH, 8DPI - PubMed Abstract:
RNA editing is a process by which post-transcriptional changes of mRNA nucleotides alter protein function through modification of the amino acid content. The 5HT 2C serotonin receptor, which undergoes 32 distinct RNA-editing events leading to 24 protein isoforms, is a notable example of this process. These 5HT 2C isoforms display differences in constitutive activity, agonist/inverse agonist potencies, and efficacies. To elucidate the molecular mechanisms responsible for these effects of RNA editing, we present four active-state 5HT 2C -transducer-coupled structures of three representative isoforms (INI, VGV, and VSV) with the selective drug lorcaserin (Belviq) and the classic psychedelic psilocin. We also provide a comprehensive analysis of agonist activation and constitutive activity across all 24 protein isoforms. Collectively, these findings reveal a unique hydrogen-bonding network located on intracellular loop 2 that is subject to RNA editing, which differentially affects GPCR constitutive and agonist signaling activities.
Organizational Affiliation:
Department of Pharmacology, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, NC 27599, USA. Electronic address: rgumpper@email.unc.edu.