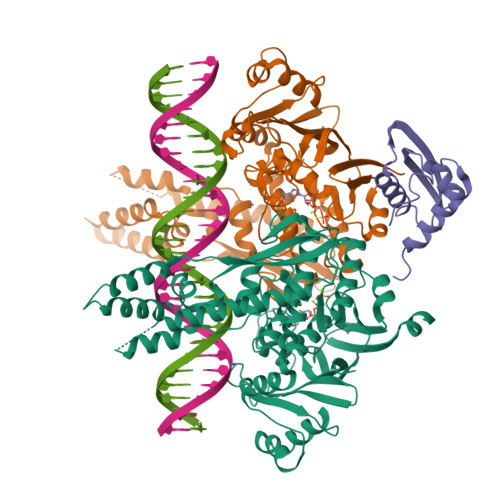
The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Deep, A., Gu, Y., Gao, Y.Q., Ego, K.M., Herzik Jr., M.A., Zhou, H., Corbett, K.D.(2022) Mol Cell 82: 4145
- PubMed: 36206765
- DOI: https://doi.org/10.1016/j.molcel.2022.09.008
- Primary Citation of Related Structures:
7TIL, 8DK1, 8DK2, 8DK3 - PubMed Abstract:
Self versus non-self discrimination is a key element of innate and adaptive immunity across life. In bacteria, CRISPR-Cas and restriction-modification systems recognize non-self nucleic acids through their sequence and their methylation state, respectively. Here, we show that the Wadjet defense system recognizes DNA topology to protect its host against plasmid transformation. By combining cryoelectron microscopy with cross-linking mass spectrometry, we show that Wadjet forms a complex similar to the bacterial condensin complex MukBEF, with a novel nuclease subunit similar to a type II DNA topoisomerase. Wadjet specifically cleaves closed-circular DNA in a reaction requiring ATP hydrolysis by the structural maintenance of chromosome (SMC) ATPase subunit JetC, suggesting that the complex could use DNA loop extrusion to sense its substrate's topology, then specifically activate the nuclease subunit JetD to cleave plasmid DNA. Overall, our data reveal how bacteria have co-opted a DNA maintenance machine to specifically recognize and destroy foreign DNAs through topology sensing.
Organizational Affiliation:
Department of Cellular and Molecular Medicine, University of California, San Diego, La Jolla, CA 92093, USA.