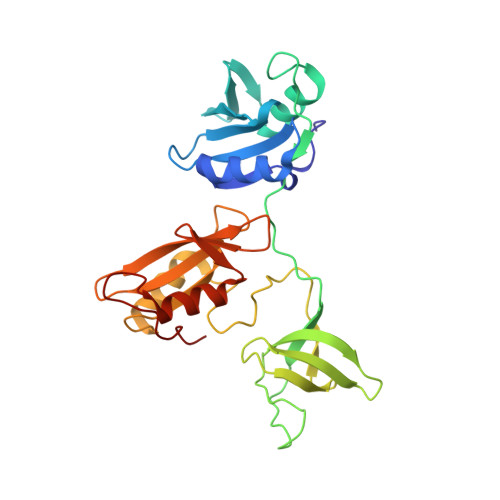
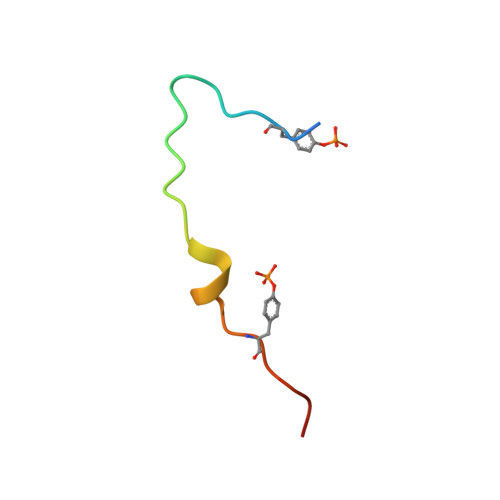
Tandem engagement of phosphotyrosines by the dual SH2 domains of p120RasGAP.
Stiegler, A.L., Vish, K.J., Boggon, T.J.(2022) Structure 30: 1603-1614.e5
- PubMed: 36417908
- DOI: https://doi.org/10.1016/j.str.2022.10.009
- Primary Citation of Related Structures:
8DGQ - PubMed Abstract:
p120RasGAP is a multidomain GTPase-activating protein for Ras. The presence of two Src homology 2 domains in an SH2-SH3-SH2 module raises the possibility that p120RasGAP simultaneously binds dual phosphotyrosine residues in target proteins. One known binding partner with two proximal phosphotyrosines is p190RhoGAP, a GTPase-activating protein for Rho GTPases. Here, we present the crystal structure of the p120RasGAP SH2-SH3-SH2 module bound to a doubly tyrosine-phosphorylated p190RhoGAP peptide, revealing simultaneous phosphotyrosine recognition by the SH2 domains. The compact arrangement places the SH2 domains in close proximity resembling an SH2 domain tandem and exposed SH3 domain. Affinity measurements support synergistic binding, while solution scattering reveals that dual phosphotyrosine binding induces compaction of this region. Our studies reflect a binding mode that limits conformational flexibility within the SH2-SH3-SH2 cassette and relies on the spacing and sequence surrounding the two phosphotyrosines, potentially representing a selectivity mechanism for downstream signaling events.
Organizational Affiliation:
Department of Pharmacology, Yale University, New Haven, CT, USA.