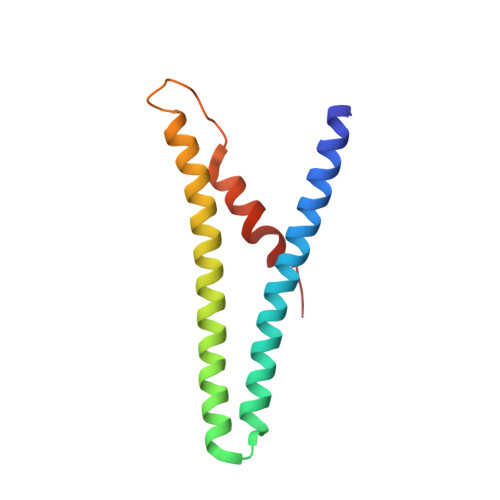
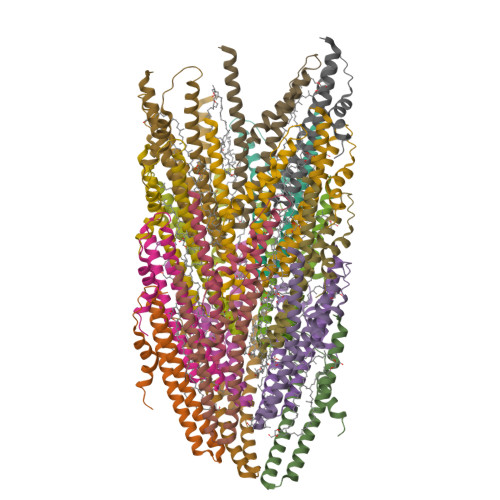Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery.
Beltran, L.C., Cvirkaite-Krupovic, V., Miller, J., Wang, F., Kreutzberger, M.A.B., Patkowski, J.B., Costa, T.R.D., Schouten, S., Levental, I., Conticello, V.P., Egelman, E.H., Krupovic, M.(2023) Nat Commun 14: 666-666
- PubMed: 36750723
- DOI: https://doi.org/10.1038/s41467-023-36349-8
- Primary Citation of Related Structures:
8DFT, 8DFU, 8EXH - PubMed Abstract:
Conjugation is a major mechanism of horizontal gene transfer promoting the spread of antibiotic resistance among human pathogens. It involves establishing a junction between a donor and a recipient cell via an extracellular appendage known as the mating pilus. In bacteria, the conjugation machinery is encoded by plasmids or transposons and typically mediates the transfer of cognate mobile genetic elements. Much less is known about conjugation in archaea. Here, we determine atomic structures by cryo-electron microscopy of three conjugative pili, two from hyperthermophilic archaea (Aeropyrum pernix and Pyrobaculum calidifontis) and one encoded by the Ti plasmid of the bacterium Agrobacterium tumefaciens, and show that the archaeal pili are homologous to bacterial mating pili. However, the archaeal conjugation machinery, known as Ced, has been 'domesticated', that is, the genes for the conjugation machinery are encoded on the chromosome rather than on mobile genetic elements, and mediates the transfer of cellular DNA.
Organizational Affiliation:
Department of Biochemistry and Molecular Genetics, University of Virginia, Charlottesville, VA, 22903, USA.