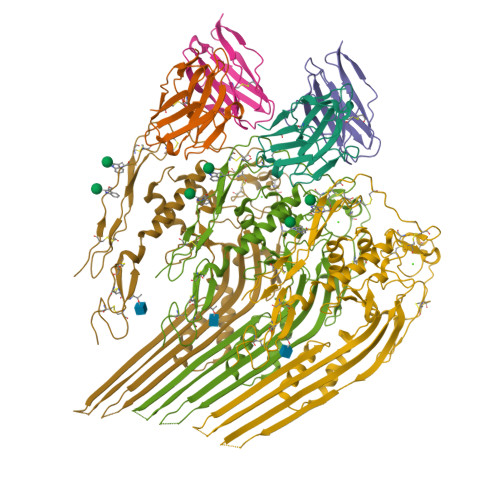
The neoepitope of the complement C5b-9 Membrane Attack Complex is formed by proximity of adjacent ancillary regions of C9.
Bayly-Jones, C., Ho, B.H.T., Lau, C., Leung, E.W.W., D'Andrea, L., Lupton, C.J., Ekkel, S.M., Venugopal, H., Whisstock, J.C., Mollnes, T.E., Spicer, B.A., Dunstone, M.A.(2023) Commun Biol 6: 42-42
- PubMed: 36639734
- DOI: https://doi.org/10.1038/s42003-023-04431-y
- Primary Citation of Related Structures:
8DE6 - PubMed Abstract:
The Membrane Attack Complex (MAC) is responsible for forming large β-barrel channels in the membranes of pathogens, such as gram-negative bacteria. Off-target MAC assembly on endogenous tissue is associated with inflammatory diseases and cancer. Accordingly, a human C5b-9 specific antibody, aE11, has been developed that detects a neoepitope exposed in C9 when it is incorporated into the C5b-9 complex, but not present in the plasma native C9. For nearly four decades aE11 has been routinely used to study complement, MAC-related inflammation, and pathophysiology. However, the identity of C9 neoepitope remains unknown. Here, we determined the cryo-EM structure of aE11 in complex with polyC9 at 3.2 Å resolution. The aE11 binding site is formed by two separate surfaces of the oligomeric C9 periphery and is therefore a discontinuous quaternary epitope. These surfaces are contributed by portions of the adjacent TSP1, LDLRA, and MACPF domains of two neighbouring C9 protomers. By substituting key antibody interacting residues to the murine orthologue, we validated the unusual binding modality of aE11. Furthermore, aE11 can recognise a partial epitope in purified monomeric C9 in vitro, albeit weakly. Taken together, our results reveal the structural basis for MAC recognition by aE11.
Organizational Affiliation:
Biomedicine Discovery Institute, Department of Biochemistry and Molecular Biology, Monash University, Melbourne, VIC, Australia.