Lipid flipping in the omega-3 fatty-acid transporter.
Nguyen, C., Lei, H.T., Lai, L.T.F., Gallenito, M.J., Mu, X., Matthies, D., Gonen, T.(2023) Nat Commun 14: 2571-2571
- PubMed: 37156797
- DOI: https://doi.org/10.1038/s41467-023-37702-7
- Primary Citation of Related Structures:
8D2S, 8D2T, 8D2U, 8D2V, 8D2W, 8D2X - PubMed Abstract:
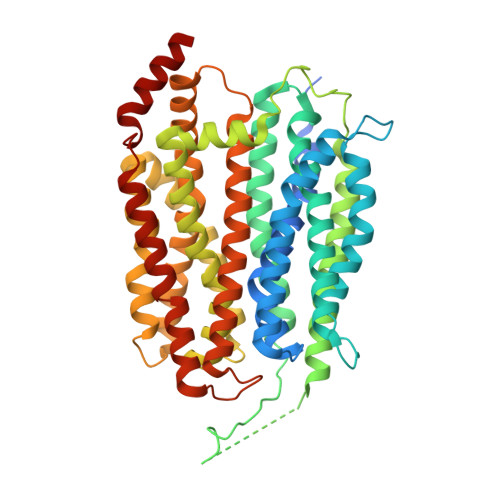
Mfsd2a is the transporter for docosahexaenoic acid (DHA), an omega-3 fatty acid, across the blood brain barrier (BBB). Defects in Mfsd2a are linked to ailments from behavioral and motor dysfunctions to microcephaly. Mfsd2a transports long-chain unsaturated fatty-acids, including DHA and α-linolenic acid (ALA), that are attached to the zwitterionic lysophosphatidylcholine (LPC) headgroup. Even with the recently determined structures of Mfsd2a, the molecular details of how this transporter performs the energetically unfavorable task of translocating and flipping lysolipids across the lipid bilayer remains unclear. Here, we report five single-particle cryo-EM structures of Danio rerio Mfsd2a (drMfsd2a): in the inward-open conformation in the ligand-free state and displaying lipid-like densities modeled as ALA-LPC at four distinct positions. These Mfsd2a snapshots detail the flipping mechanism for lipid-LPC from outer to inner membrane leaflet and release for membrane integration on the cytoplasmic side. These results also map Mfsd2a mutants that disrupt lipid-LPC transport and are associated with disease.
Organizational Affiliation:
Howard Hughes Medical Institute, University of California Los Angeles, Los Angeles, CA, 90095, USA.