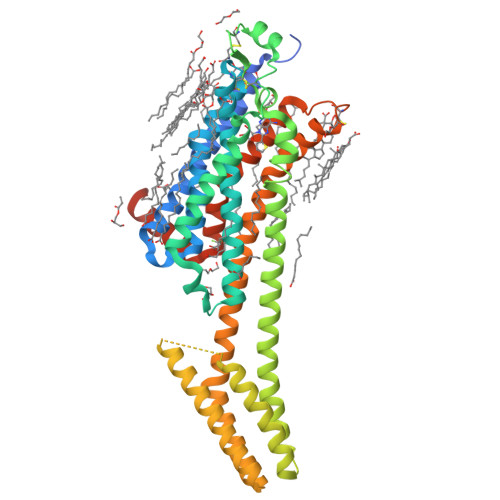
GPCR Agonist-to-Antagonist Conversion: Enabling the Design of Nucleoside Functional Switches for the A 2A Adenosine Receptor.
Shiriaeva, A., Park, D., Kim, G., Lee, Y., Hou, X., Jarhad, D.B., Kim, G., Yu, J., Hyun, Y.E., Kim, W., Gao, Z.G., Jacobson, K.A., Han, G.W., Stevens, R.C., Jeong, L.S., Choi, S., Cherezov, V.(2022) J Med Chem 65: 11648-11657
- PubMed: 35977382
- DOI: https://doi.org/10.1021/acs.jmedchem.2c00462
- Primary Citation of Related Structures:
8CU6, 8CU7 - PubMed Abstract:
Modulators of the G protein-coupled A 2A adenosine receptor (A 2A AR) have been considered promising agents to treat Parkinson's disease, inflammation, cancer, and central nervous system disorders. Herein, we demonstrate that a thiophene modification at the C8 position in the common adenine scaffold converted an A 2A AR agonist into an antagonist. We synthesized and characterized a novel A 2A AR antagonist, 2 (LJ-4517), with K i = 18.3 nM. X-ray crystallographic structures of 2 in complex with two thermostabilized A 2A AR constructs were solved at 2.05 and 2.80 Å resolutions. In contrast to A 2A AR agonists, which simultaneously interact with both Ser277 7.42 and His278 7.43 , 2 only transiently contacts His278 7.43 , which can be direct or water-mediated. The n -hexynyl group of 2 extends into an A 2A AR exosite. Structural analysis revealed that the introduced thiophene modification restricted receptor conformational rearrangements required for subsequent activation. This approach can expand the repertoire of adenosine receptor antagonists that can be designed based on available agonist scaffolds.
Organizational Affiliation:
Department of Chemistry, University of Southern California, Los Angeles, California 90089, United States.