Potent cross-reactive mAbs from BA.4/5 breakthrough infection
Duyvesteyn, H.M.E., Screaton, G., Ren, J., Stuart, D.I., Fry, E.E.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Spike protein S2' | A [auth E] | 202 | Homo sapiens | Mutation(s): 0 Gene Names: S, 2 | 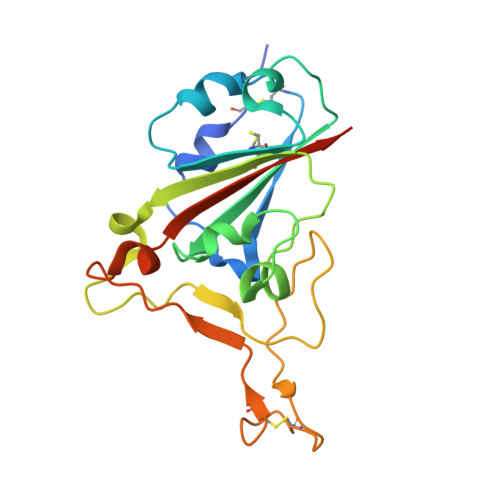 |
UniProt | |||||
Find proteins for P0DTC2 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTC2 Go to UniProtKB: P0DTC2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTC2 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | Go to GlyGen: P0DTC2-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| SARS1-34 fab Heavy Chain | B [auth H] | 227 | Homo sapiens | Mutation(s): 0 | 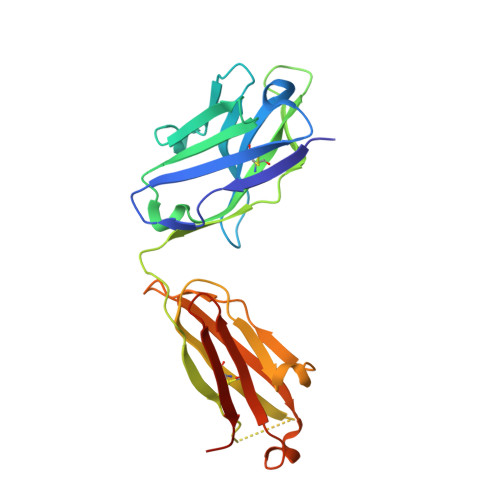 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| SARS1-34 fab Light Chain | C [auth L] | 214 | Homo sapiens | Mutation(s): 0 | 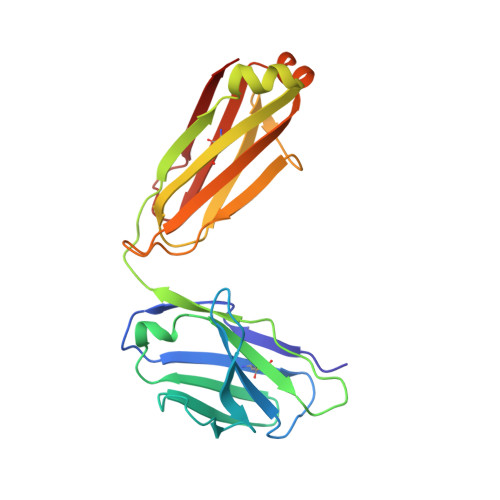 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| BA.2-07 fab Heavy Chain | D [auth A] | 231 | Homo sapiens | Mutation(s): 0 | 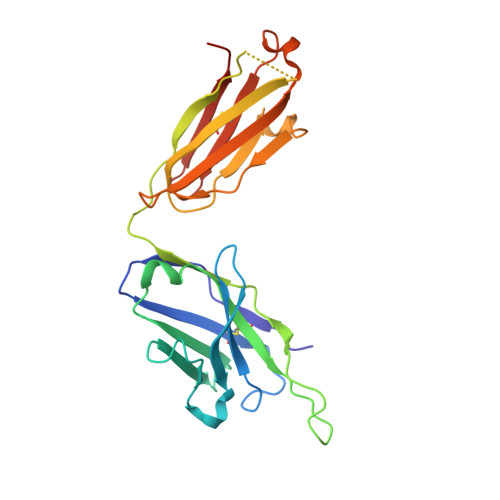 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| BA.2-07 fab Light Chain | E [auth B] | 214 | Homo sapiens | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| C1 nanobody | F [auth C] | 131 | Lama glama | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | G [auth D] | 4 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G32152BH GlyCosmos: G32152BH GlyGen: G32152BH | |||||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Wellcome Trust | United Kingdom | 203141/A/16/Z |
| Wellcome Trust | United Kingdom | 101122/Z/13/Z |
| Chinese Academy of Sciences | China | 2018-I2M-2-002 |
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MR/N00065X/1 |