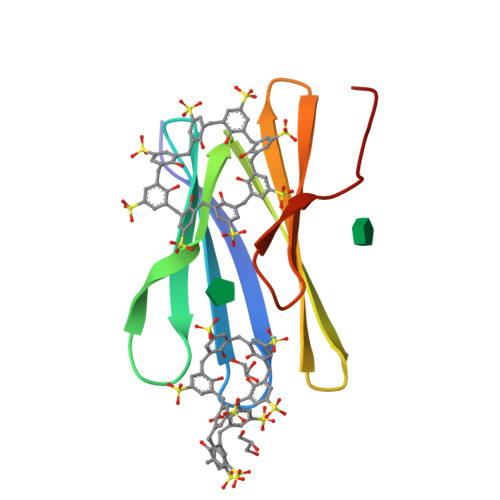
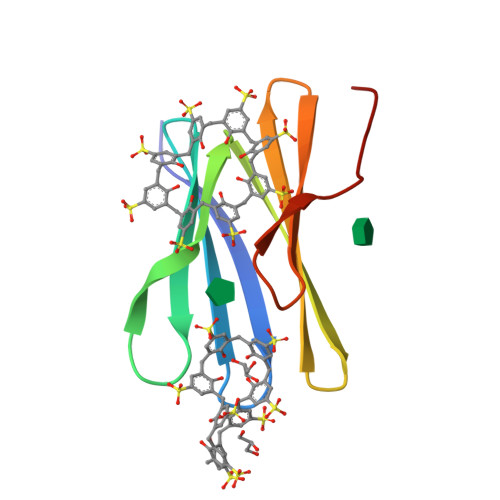
Protein-macrocycle polymorphism: crystal form IV of the Ralstonia solanacearum lectin-sulfonato-calix[8]arene complex.
Mockler, N.M., Ramberg, K.O., Crowley, P.B.(2023) Acta Crystallogr D Struct Biol 79: 624-631
- PubMed: 37314405
- DOI: https://doi.org/10.1107/S2059798323003832
- Primary Citation of Related Structures:
8C9Y, 8C9Z - PubMed Abstract:
Controlled protein assembly and crystallization is necessary as a means of generating diffraction-quality crystals as well as providing a basis for new types of biomaterials. Water-soluble calixarenes are useful mediators of protein crystallization. Recently, it was demonstrated that Ralstonia solanacearum lectin (RSL) co-crystallizes with anionic sulfonato-calix[8]arene (sclx 8 ) in three space groups. Two of these co-crystals only grow at pH ≤ 4 where the protein is cationic, and the crystal packing is dominated by the calixarene. This paper describes a fourth RSL-sclx 8 co-crystal, which was discovered while working with a cation-enriched mutant. Crystal form IV grows at high ionic strength in the pH range 5-6. While possessing some features in common with the previous forms, the new structure reveals alternative calixarene binding modes. The occurrence of C 2 -symmetric assemblies, with the calixarene at special positions, appears to be an important result for framework fabrication. Questions arise regarding crystal screening and exhaustive searching for polymorphs.
Organizational Affiliation:
School of Biological and Chemical Sciences, University of Galway, University Road, Galway H91 TK33, Ireland.