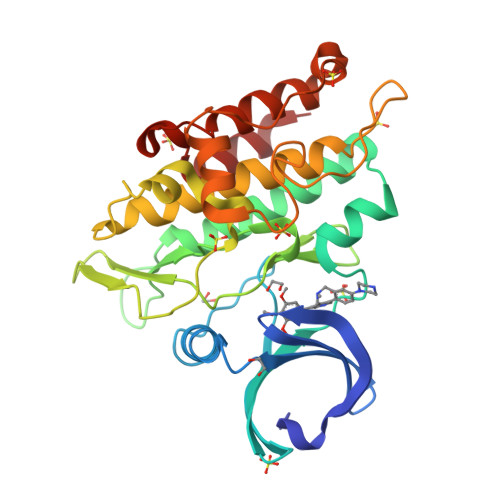
Discovery of Conformationally Constrained ALK2 Inhibitors.
Gonzalez-Alvarez, H., Ensan, D., Xin, T., Wong, J.F., Zepeda-Velazquez, C.A., Cros, J., Sweeney, M.N., Hoffer, L., Kiyota, T., Wilson, B.J., Aman, A., Roberts, O., Isaac, M.B., Bullock, A.N., Smil, D., Al-Awar, R.(2024) J Med Chem 67: 4707-4725
- PubMed: 38498998
- DOI: https://doi.org/10.1021/acs.jmedchem.3c02308
- Primary Citation of Related Structures:
8C7W, 8C7Z - PubMed Abstract:
Despite decades of research on new diffuse intrinsic pontine glioma (DIPG) treatments, little or no progress has been made on improving patient outcomes. In this work, we explored novel scaffold modifications of M4K2009 , a 3,5-diphenylpyridine ALK2 inhibitor previously reported by our group. Here we disclose the design, synthesis, and evaluation of a first-in-class set of 5- to 7-membered ether-linked and 7-membered amine-linked constrained inhibitors of ALK2. This rigidification strategy led us to the discovery of the ether-linked inhibitors M4K2308 and M4K2281 and the amine-linked inhibitors M4K2304 and M4K2306 , each with superior potency against ALK2. Notably, M4K2304 and M4K2306 exhibit exceptional selectivity for ALK2 over ALK5, surpassing the reference compound. Preliminary studies on their in vivo pharmacokinetics, including blood-brain barrier penetration, revealed that these constrained scaffolds have favorable exposure and do open a novel chemical space for further optimization and future evaluation in orthotopic models of DIPG.
Organizational Affiliation:
Drug Discovery Program, Ontario Institute for Cancer Research, 661 University Avenue, MaRS Centre, West Tower, Toronto, Ontario M5G 0A3, Canada.