Immunization with lytic polysaccharide monooxygenase CbpD induces protective immunity against Pseudomonas aeruginosa pneumonia.
Askarian, F., Tsai, C.M., Cordara, G., Zurich, R.H., Bjanes, E., Golten, O., Vinther Sorensen, H., Kousha, A., Meier, A., Chikwati, E., Bruun, J.A., Ludviksen, J.A., Choudhury, B., Trieu, D., Davis, S., Edvardsen, P.K.T., Mollnes, T.E., Liu, G.Y., Krengel, U., Conrad, D.J., Vaaje-Kolstad, G., Nizet, V.(2023) Proc Natl Acad Sci U S A 120: e2301538120-e2301538120
- PubMed: 37459522
- DOI: https://doi.org/10.1073/pnas.2301538120
- Primary Citation of Related Structures:
8C5N - PubMed Abstract:
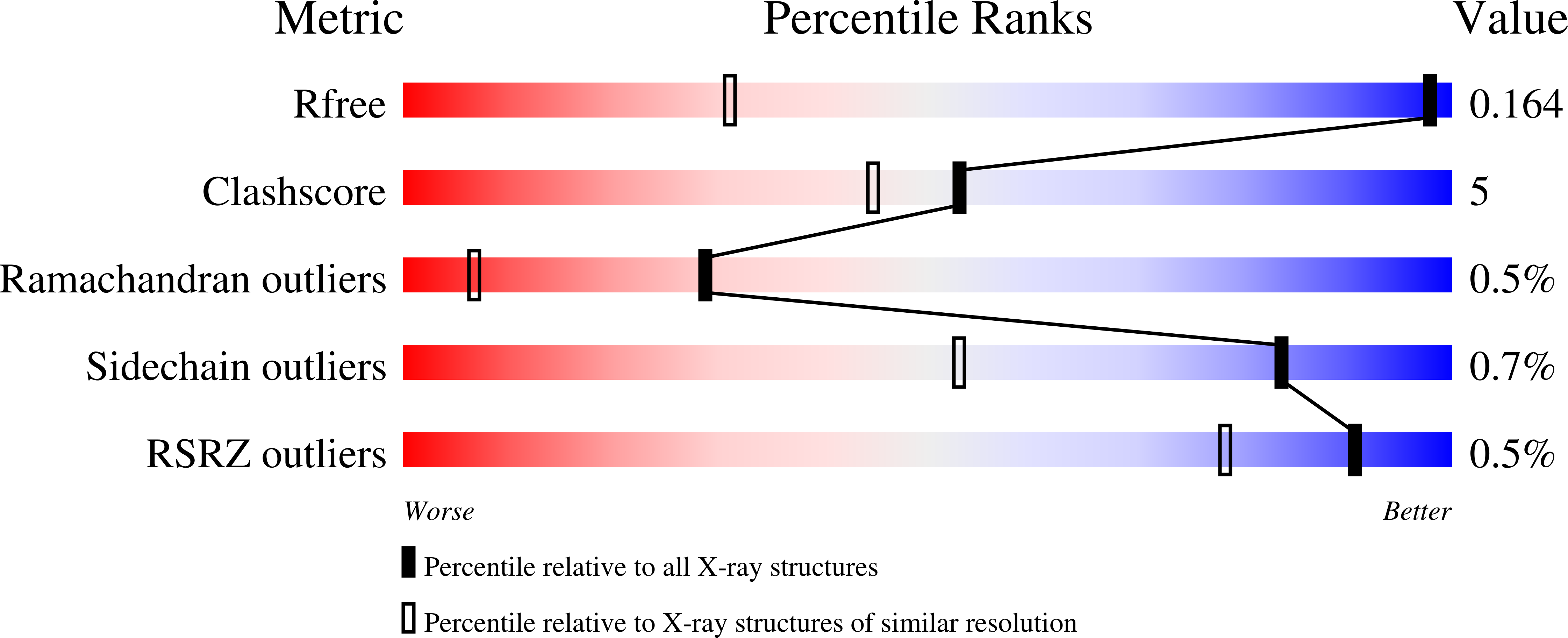
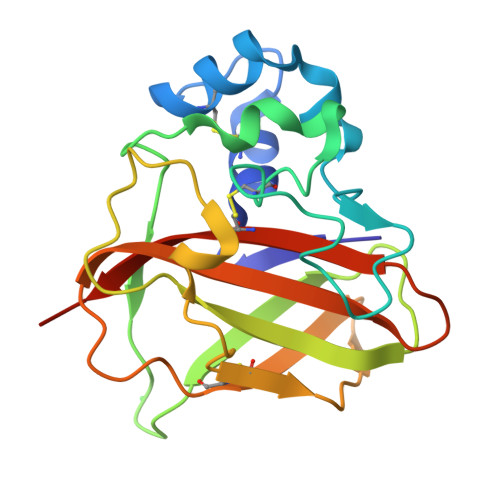
Pseudomonas aeruginosa (PA) CbpD belongs to the lytic polysaccharide monooxygenases (LPMOs), a family of enzymes that cleave chitin or related polysaccharides. Here, we demonstrate a virulence role of CbpD in PA pneumonia linked to impairment of host complement function and opsonophagocytic clearance. Following intratracheal challenge, a PA ΔCbpD mutant was more easily cleared and produced less mortality than the wild-type parent strain. The x-ray crystal structure of the CbpD LPMO domain was solved to subatomic resolution (0.75Å) and its two additional domains modeled by small-angle X-ray scattering and Alphafold2 machine-learning algorithms, allowing structure-based immune epitope mapping. Immunization of naive mice with recombinant CbpD generated high IgG antibody titers that promoted human neutrophil opsonophagocytic killing, neutralized enzymatic activity, and protected against lethal PA pneumonia and sepsis. IgG antibodies generated against full-length CbpD or its noncatalytic M2+CBM73 domains were opsonic and protective, even in previously PA-exposed mice, while antibodies targeting the AA10 domain were not. Preexisting antibodies in PA-colonized cystic fibrosis patients primarily target the CbpD AA10 catalytic domain. Further exploration of LPMO family proteins, present across many clinically important and antibiotic-resistant human pathogens, may yield novel and effective vaccine antigens.
Organizational Affiliation:
Division of Host-Microbe Systems & Therapeutics, Department of Pediatrics, University of California San Diego, La Jolla, CA 92093.