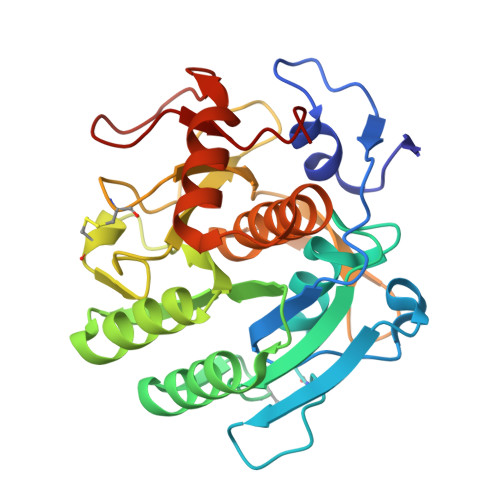
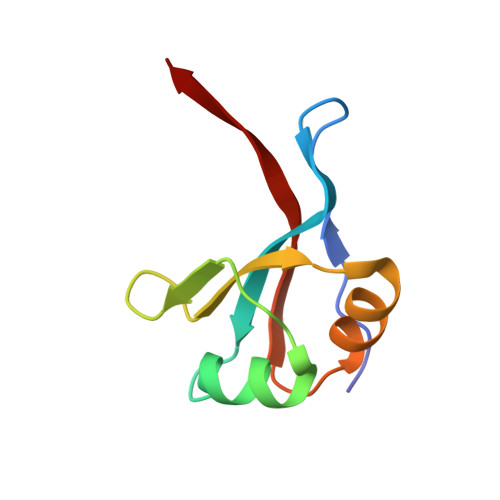
An engineered enzyme embedded into PLA to make self-biodegradable plastic.
Guicherd, M., Ben Khaled, M., Gueroult, M., Nomme, J., Dalibey, M., Grimaud, F., Alvarez, P., Kamionka, E., Gavalda, S., Noel, M., Vuillemin, M., Amillastre, E., Labourdette, D., Cioci, G., Tournier, V., Kitpreechavanich, V., Dubois, P., Andre, I., Duquesne, S., Marty, A.(2024) Nature 631: 884-890
- PubMed: 39020178
- DOI: https://doi.org/10.1038/s41586-024-07709-1
- Primary Citation of Related Structures:
8C4X, 8C4Z - PubMed Abstract:
Plastic production reached 400 million tons in 2022 (ref. 1 ), with packaging and single-use plastics accounting for a substantial amount of this 2 . The resulting waste ends up in landfills, incineration or the environment, contributing to environmental pollution 3 . Shifting to biodegradable and compostable plastics is increasingly being considered as an efficient waste-management alternative 4 . Although polylactide (PLA) is the most widely used biosourced polymer 5 , its biodegradation rate under home-compost and soil conditions remains low 6-8 . Here we present a PLA-based plastic in which an optimized enzyme is embedded to ensure rapid biodegradation and compostability at room temperature, using a scalable industrial process. First, an 80-fold activity enhancement was achieved through structure-based rational engineering of a new hyperthermostable PLA hydrolase. Second, the enzyme was uniformly dispersed within the PLA matrix by means of a masterbatch-based melt extrusion process. The liquid enzyme formulation was incorporated in polycaprolactone, a low-melting-temperature polymer, through melt extrusion at 70 °C, forming an 'enzymated' polycaprolactone masterbatch. Masterbatch pellets were integrated into PLA by melt extrusion at 160 °C, producing an enzymated PLA film (0.02% w/w enzyme) that fully disintegrated under home-compost conditions within 20-24 weeks, meeting home-composting standards. The mechanical and degradation properties of the enzymated film were compatible with industrial packaging applications, and they remained intact during long-term storage. This innovative material not only opens new avenues for composters and biomethane production but also provides a feasible industrial solution for PLA degradation.
Organizational Affiliation:
Toulouse Biotechnology Institute, Université de Toulouse, CNRS, INRAE, INSA, Toulouse, France.