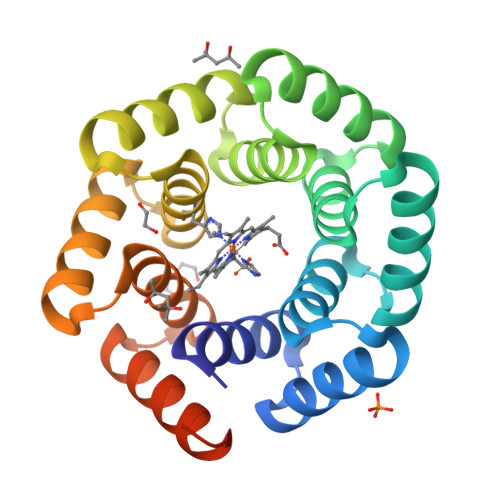
Design of Heme Enzymes with a Tunable Substrate Binding Pocket Adjacent to an Open Metal Coordination Site.
Kalvet, I., Ortmayer, M., Zhao, J., Crawshaw, R., Ennist, N.M., Levy, C., Roy, A., Green, A.P., Baker, D.(2023) J Am Chem Soc 145: 14307-14315
- PubMed: 37341421
- DOI: https://doi.org/10.1021/jacs.3c02742
- Primary Citation of Related Structures:
8C3W - PubMed Abstract:
The catalytic versatility of pentacoordinated iron is highlighted by the broad range of natural and engineered activities of heme enzymes such as cytochrome P450s, which position a porphyrin cofactor coordinating a central iron atom below an open substrate binding pocket. This catalytic prowess has inspired efforts to design de novo helical bundle scaffolds that bind porphyrin cofactors. However, such designs lack the large open substrate binding pocket of P450s, and hence, the range of chemical transformations accessible is limited. Here, with the goal of combining the advantages of the P450 catalytic site geometry with the almost unlimited customizability of de novo protein design, we design a high-affinity heme-binding protein, dnHEM1, with an axial histidine ligand, a vacant coordination site for generating reactive intermediates, and a tunable distal pocket for substrate binding. A 1.6 Å X-ray crystal structure of dnHEM1 reveals excellent agreement to the design model with key features programmed as intended. The incorporation of distal pocket substitutions converted dnHEM1 into a proficient peroxidase with a stable neutral ferryl intermediate. In parallel, dnHEM1 was redesigned to generate enantiocomplementary carbene transferases for styrene cyclopropanation (up to 93% isolated yield, 5000 turnovers, 97:3 e.r.) by reconfiguring the distal pocket to accommodate calculated transition state models. Our approach now enables the custom design of enzymes containing cofactors adjacent to binding pockets with an almost unlimited variety of shapes and functionalities.
Organizational Affiliation:
Institute for Protein Design, University of Washington, 3946 W Stevens Way NE, Seattle, Washington 98195, United States.