Properties and Crystal Structure of the Cereibacter sphaeroides Photosynthetic Reaction Center with Double Amino Acid Substitution I(L177)H + F(M197)H.
Fufina, T.Y., Selikhanov, G.K., Gabdulkhakov, A.G., Vasilieva, L.G.(2023) Membranes (Basel) 13
- PubMed: 36837660
- DOI: https://doi.org/10.3390/membranes13020157
- Primary Citation of Related Structures:
8C3F - PubMed Abstract:
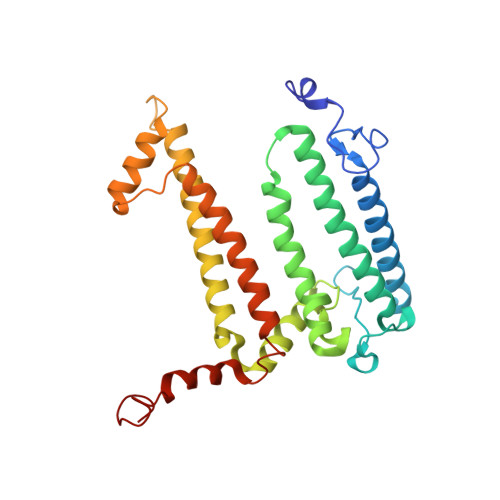
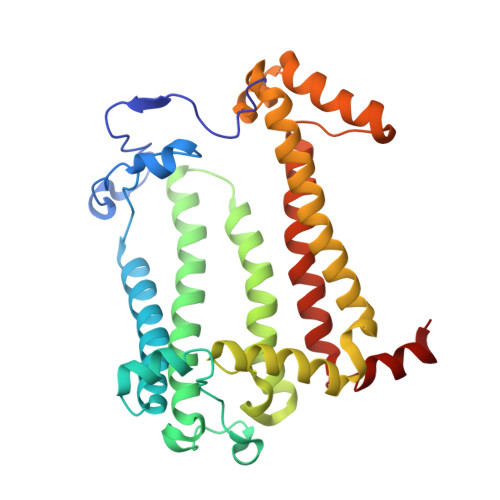
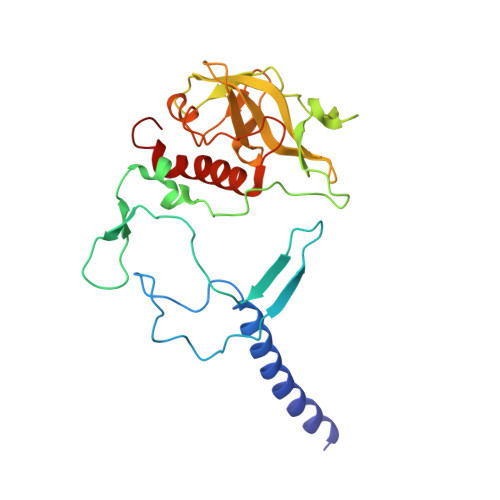
The photosynthetic reaction center of the purple bacterium Cereibacter sphaeroides with two site-directed mutations Ile-L177-His and M197 Phe-His is of double interest. The substitution I(L177)H results in strong binding of a bacteriochlorophyll molecule with L-subunit. The second mutation F(M197)H introduces a new H-bond between the C2-acetyl carbonyl group of the bacteriochlorophyll P B and His-M197, which is known to enhance the stability of the complex. Due to this H-bond, π -electron system of P finds itself connected to an extensive H-bonding network on the periplasmic surface of the complex. The crystal structure of the double mutant reaction center obtained with 2.6 Å resolution allows clarifying consequences of the Ile L177 - His substitution. The value of the P/P + midpoint potential in the double mutant RC was found to be ~20 mV less than the sum of potentials measured in the two RCs with single mutations I(L177)H and F(M197)H. The protein environment of the BChls P A and B B were found to be similar to that in the RC with single substitution I(L177)H, whereas an altered pattern of the H-bonding networks was found in the vicinity of bacteriochlorophyll P B . The data obtained are consistent with our previous assumption on a correlation between the bulk of the H-bonding network connected with the π-electron system of the primary electron donor P and the value of its oxidation potential.
Organizational Affiliation:
Federal Research Center Pushchino Scientific Center for Biological Research PSCBR, Institute of Basic Biological Problems, Russian Academy of Sciences, Institutskaya Street 2, 142290 Pushchino, Russia.