Activator-induced conformational changes regulate division-associated peptidoglycan amidases.
Cook, J., Baverstock, T.C., McAndrew, M.B.L., Roper, D.I., Stansfeld, P.J., Crow, A.(2023) Proc Natl Acad Sci U S A 120: e2302580120-e2302580120
- PubMed: 37276423
- DOI: https://doi.org/10.1073/pnas.2302580120
- Primary Citation of Related Structures:
8C0J, 8C2O - PubMed Abstract:
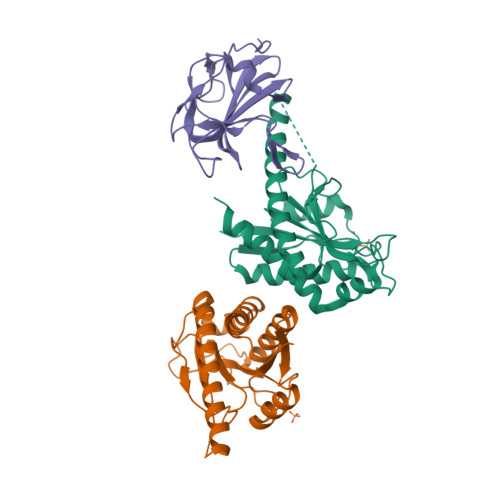
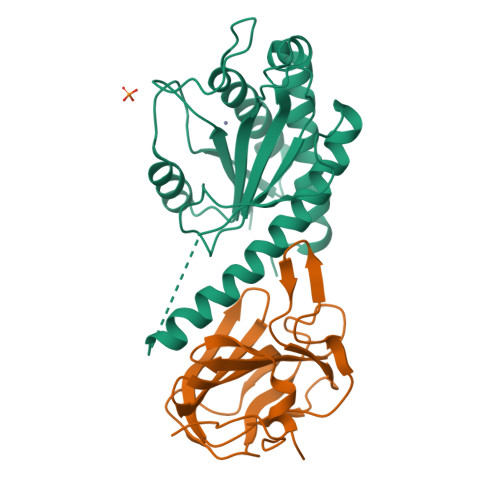
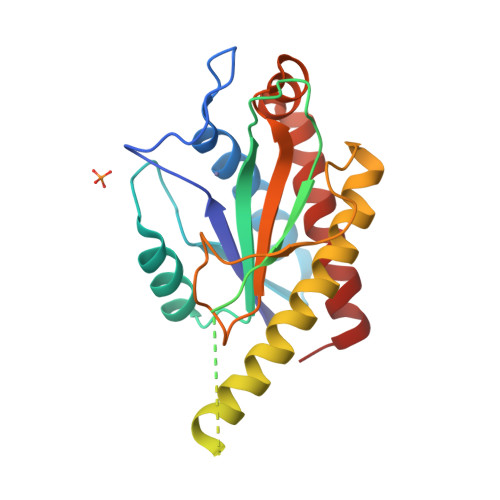
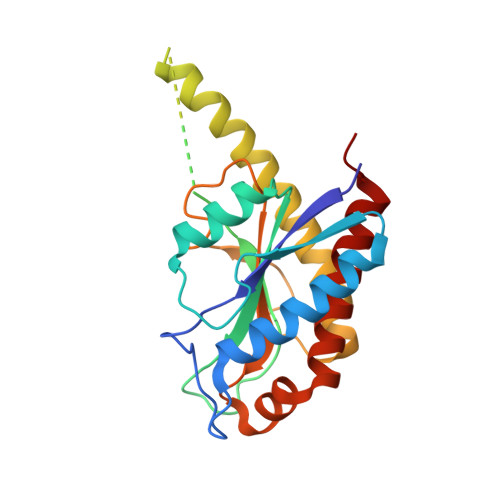
AmiA and AmiB are peptidoglycan-hydrolyzing enzymes from Escherichia coli that are required to break the peptidoglycan layer during bacterial cell division and maintain integrity of the cell envelope. In vivo, the activity of AmiA and AmiB is tightly controlled through their interactions with the membrane-bound FtsEX-EnvC complex. Activation of AmiA and AmiB requires access to a groove in the amidase-activating LytM domain of EnvC which is gated by ATP-driven conformational changes in FtsEX-EnvC complex. Here, we present a high-resolution structure of the isolated AmiA protein, confirming that it is autoinhibited in the same manner as AmiB and AmiC, and a complex of the AmiB enzymatic domain bound to the activating EnvC LytM domain. In isolation, the active site of AmiA is blocked by an autoinhibitory helix that binds directly to the catalytic zinc and fills the volume expected to accommodate peptidoglycan binding. In the complex, binding of the EnvC LytM domain induces a conformational change that displaces the amidase autoinhibitory helix and reorganizes the active site for activity. Our structures, together with complementary mutagenesis work, defines the conformational changes required to activate AmiA and/or AmiB through their interaction with their cognate activator EnvC.
Organizational Affiliation:
School of Life Sciences, University of Warwick, Coventry CV4 7AL, United Kingdom.