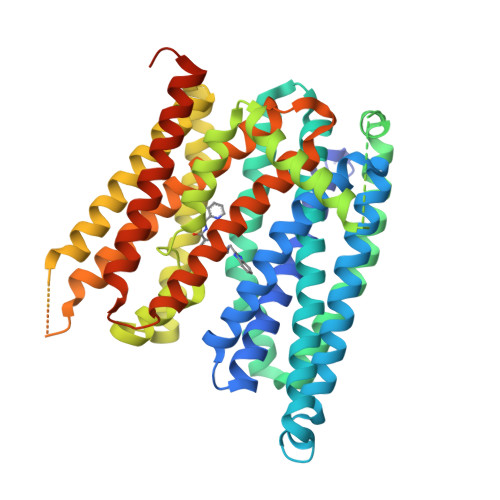
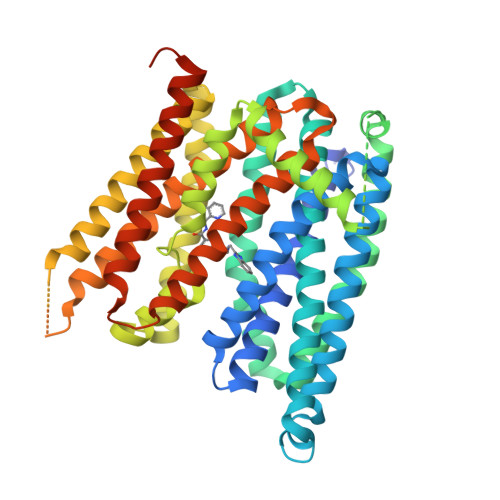
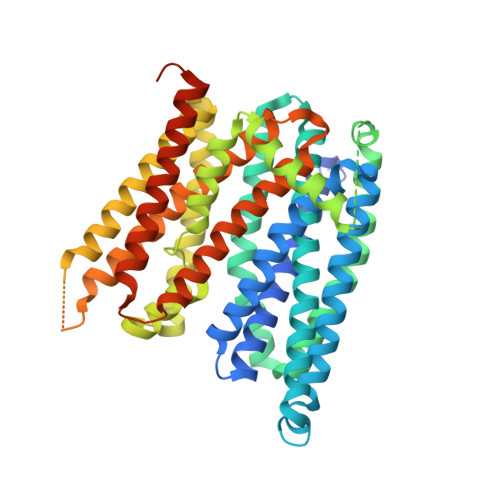
Structures of ferroportin in complex with its specific inhibitor vamifeport.
Lehmann, E.F., Liziczai, M., Drozdzyk, K., Altermatt, P., Langini, C., Manolova, V., Sundstrom, H., Durrenberger, F., Dutzler, R., Manatschal, C.(2023) Elife 12
- PubMed: 36943194
- DOI: https://doi.org/10.7554/eLife.83053
- Primary Citation of Related Structures:
8BZY, 8C02, 8C03 - PubMed Abstract:
A central regulatory mechanism of iron homeostasis in humans involves ferroportin (FPN), the sole cellular iron exporter, and the peptide hormone hepcidin, which inhibits Fe 2+ transport and induces internalization and degradation of FPN. Dysregulation of the FPN/hepcidin axis leads to diverse pathological conditions, and consequently, pharmacological compounds that inhibit FPN-mediated iron transport are of high clinical interest. Here, we describe the cryo-electron microscopy structures of human FPN in complex with synthetic nanobodies and vamifeport (VIT-2763), the first clinical-stage oral FPN inhibitor. Vamifeport competes with hepcidin for FPN binding and is currently in clinical development for β-thalassemia and sickle cell disease. The structures display two distinct conformations of FPN, representing outward-facing and occluded states of the transporter. The vamifeport site is located in the center of the protein, where the overlap with hepcidin interactions underlies the competitive relationship between the two molecules. The introduction of point mutations in the binding pocket of vamifeport reduces its affinity to FPN, emphasizing the relevance of the structural data. Together, our study reveals conformational rearrangements of FPN that are of potential relevance for transport, and it provides initial insight into the pharmacological targeting of this unique iron efflux transporter.
Organizational Affiliation:
Department of Biochemistry, University of Zurich, Zürich, Switzerland.