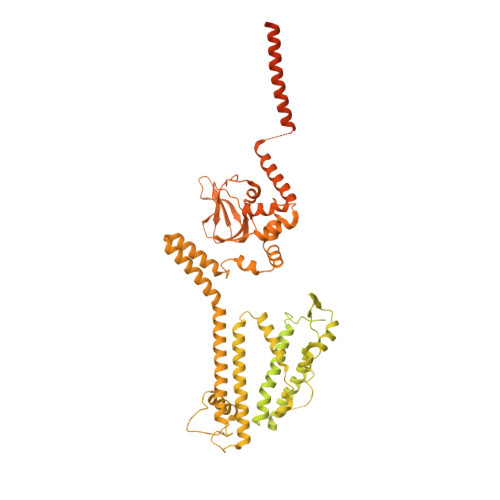
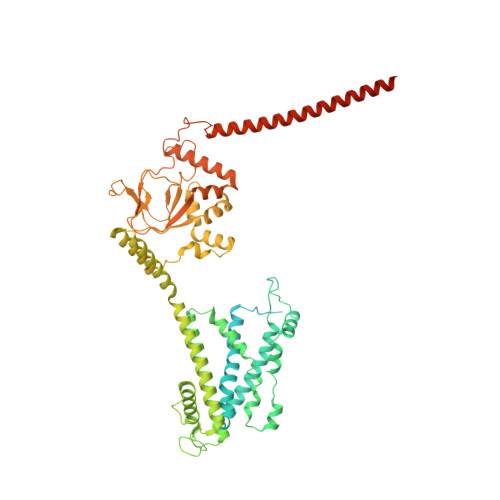
Structural basis of calmodulin modulation of the rod cyclic nucleotide-gated channel.
Barret, D.C.A., Schuster, D., Rodrigues, M.J., Leitner, A., Picotti, P., Schertler, G.F.X., Kaupp, U.B., Korkhov, V.M., Marino, J.(2023) Proc Natl Acad Sci U S A 120: e2300309120-e2300309120
- PubMed: 37011209
- DOI: https://doi.org/10.1073/pnas.2300309120
- Primary Citation of Related Structures:
8BX7 - PubMed Abstract:
Calmodulin (CaM) regulates many ion channels to control calcium entry into cells, and mutations that alter this interaction are linked to fatal diseases. The structural basis of CaM regulation remains largely unexplored. In retinal photoreceptors, CaM binds to the CNGB subunit of cyclic nucleotide-gated (CNG) channels and, thereby, adjusts the channel's Cyclic guanosine monophosphate (cGMP) sensitivity in response to changes in ambient light conditions. Here, we provide the structural characterization for CaM regulation of a CNG channel by using a combination of single-particle cryo-electron microscopy and structural proteomics. CaM connects the CNGA and CNGB subunits, resulting in structural changes both in the cytosolic and transmembrane regions of the channel. Cross-linking and limited proteolysis-coupled mass spectrometry mapped the conformational changes induced by CaM in vitro and in the native membrane. We propose that CaM is a constitutive subunit of the rod channel to ensure high sensitivity in dim light. Our mass spectrometry-based approach is generally relevant for studying the effect of CaM on ion channels in tissues of medical interest, where only minute quantities are available.
Organizational Affiliation:
Laboratory of Biomolecular Research, Paul Scherrer Institute, 5232 Villigen, Switzerland.