Iron-only Fe-nitrogenase underscores common catalytic principles in biological nitrogen fixation
Trncik, C., Detemple, F., Einsle, O.(2023) Nat Catal
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2023) Nat Catal
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nitrogenase protein alpha chain | 515 | Azotobacter vinelandii DJ | Mutation(s): 0 EC: 1.18.6.1 | 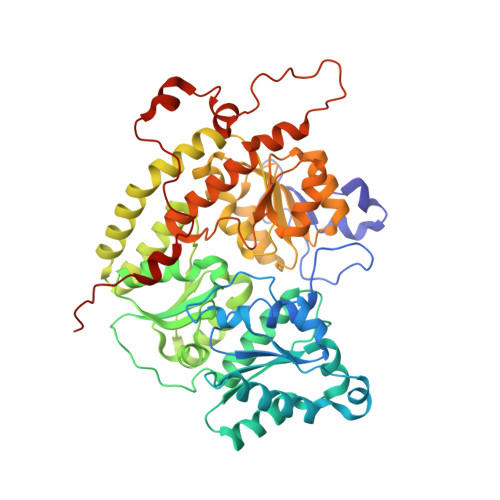 | |
UniProt | |||||
Find proteins for P16266 (Azotobacter vinelandii) Explore P16266 Go to UniProtKB: P16266 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P16266 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Fe-only nitrogenase, beta subunit | 461 | Azotobacter vinelandii DJ | Mutation(s): 0 EC: 1.18.6.1 | 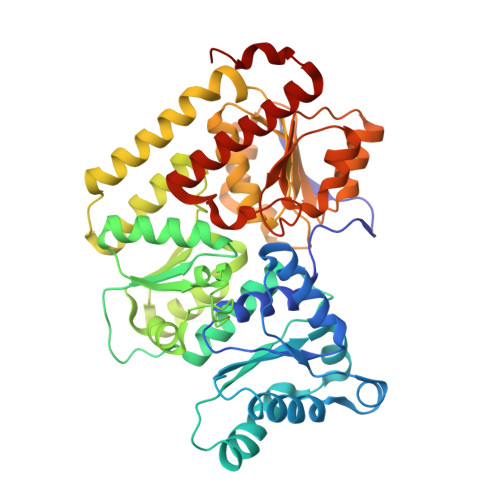 | |
UniProt | |||||
Find proteins for P16267 (Azotobacter vinelandii) Explore P16267 Go to UniProtKB: P16267 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P16267 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nitrogenase iron-iron protein delta chain | 119 | Azotobacter vinelandii DJ | Mutation(s): 0 EC: 1.18.6.1 | 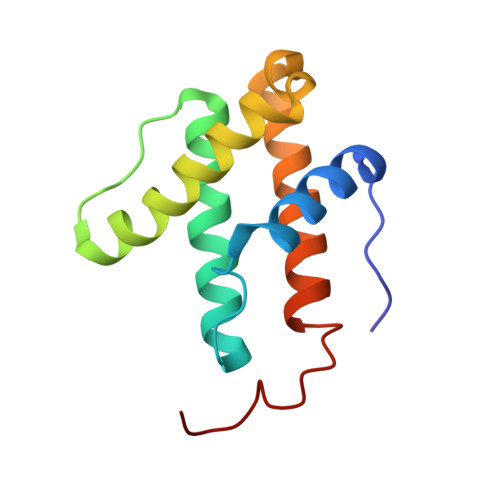 | |
UniProt | |||||
Find proteins for P16268 (Azotobacter vinelandii) Explore P16268 Go to UniProtKB: P16268 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P16268 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| S5Q (Subject of Investigation/LOI) Query on S5Q | G [auth A], N [auth D] | FeFe cofactor C Fe8 S9 ARHQIUGQOSHZFD-UHFFFAOYSA-N |  | ||
| CLF Query on CLF | I [auth A], R [auth E] | FE(8)-S(7) CLUSTER Fe8 S7 JKVMXLBGZBULKV-UHFFFAOYSA-N |  | ||
| HCA Query on HCA | H [auth A], O [auth D] | 3-HYDROXY-3-CARBOXY-ADIPIC ACID C7 H10 O7 XKJVEVRQMLKSMO-SSDOTTSWSA-N |  | ||
| H2S Query on H2S | K [auth A], Q [auth D] | HYDROSULFURIC ACID H2 S RWSOTUBLDIXVET-UHFFFAOYSA-N |  | ||
| MG Query on MG | L [auth B], M [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| O Query on O | J [auth A], P [auth D] | OXYGEN ATOM O XLYOFNOQVPJJNP-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 109.969 | α = 90 |
| b = 151.048 | β = 90 |
| c = 158.863 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| REFMAC | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| MOLREP | phasing |
| Coot | model building |
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Research Foundation (DFG) | Germany | PP 1927, project ID 311061829 |
| German Research Foundation (DFG) | Germany | RTG 1976, project ID 235777276 |
| European Research Council (ERC) | European Union | 310676 |