Distant sequence regions of JBP1 contribute to J-DNA binding.
de Vries, I., Ammerlaan, D., Heidebrecht, T., Celie, P.H., Geerke, D.P., Joosten, R.P., Perrakis, A.(2023) Life Sci Alliance 6
- PubMed: 37328191
- DOI: https://doi.org/10.26508/lsa.202302150
- Primary Citation of Related Structures:
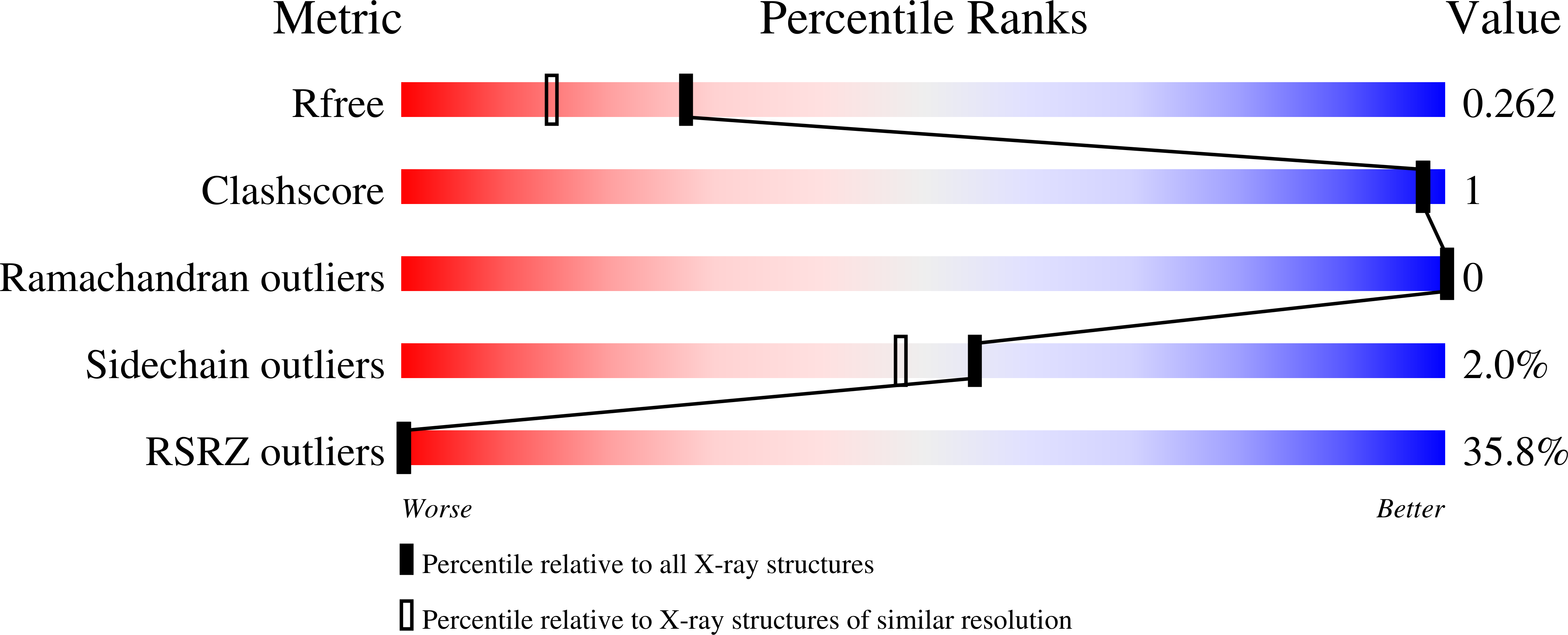
8BBM - PubMed Abstract:
Base-J (β-D-glucopyranosyloxymethyluracil) is a modified DNA nucleotide that replaces 1% of thymine in kinetoplastid flagellates. The biosynthesis and maintenance of base-J depends on the base-J-binding protein 1 (JBP1) that has a thymidine hydroxylase domain and a J-DNA-binding domain (JDBD). How the thymidine hydroxylase domain synergizes with the JDBD to hydroxylate thymine in specific genomic sites, maintaining base-J during semi-conservative DNA replication, remains unclear. Here, we present a crystal structure of the JDBD including a previously disordered DNA-contacting loop and use it as starting point for molecular dynamics simulations and computational docking studies to propose recognition models for JDBD binding to J-DNA. These models guided mutagenesis experiments, providing additional data for docking, which reveals a binding mode for JDBD onto J-DNA. This model, together with the crystallographic structure of the TET2 JBP1-homologue in complex with DNA and the AlphaFold model of full-length JBP1, allowed us to hypothesize that the flexible JBP1 N-terminus contributes to DNA-binding, which we confirmed experimentally. Α high-resolution JBP1:J-DNA complex, which must involve conformational changes, would however need to be determined experimentally to further understand this unique underlying molecular mechanism that ensures replication of epigenetic information.
Organizational Affiliation:
Oncode Institute and Division of Biochemistry, Netherlands Cancer Institute, Amsterdam, The Netherlands.