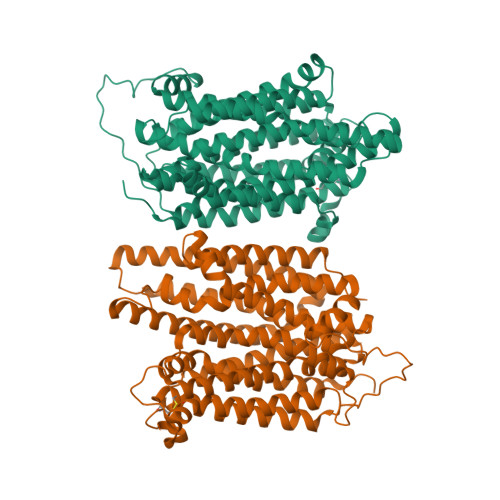
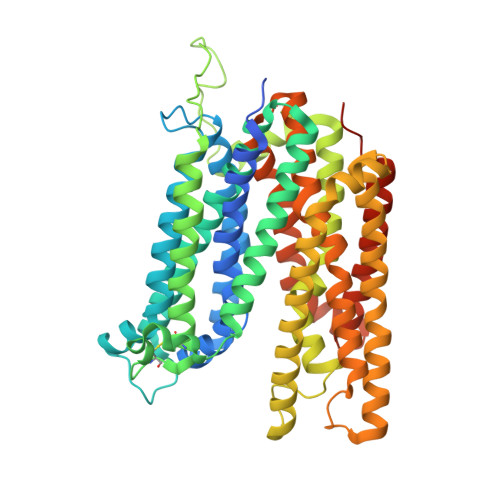
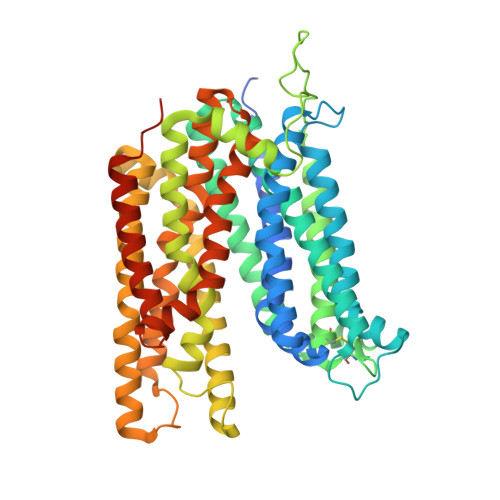
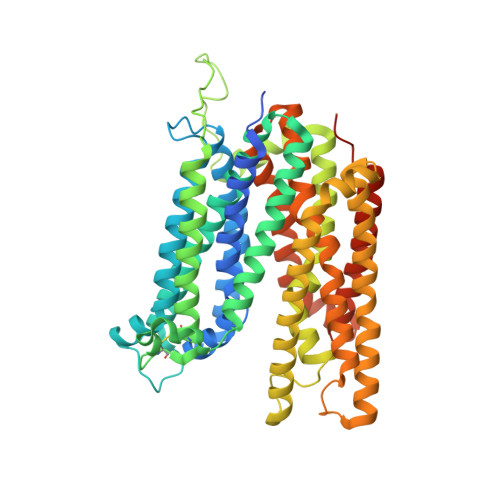
Structure and sucrose binding mechanism of the plant SUC1 sucrose transporter.
Bavnhoj, L., Driller, J.H., Zuzic, L., Stange, A.D., Schiott, B., Pedersen, B.P.(2023) Nat Plants 9: 938-950
- PubMed: 37188854
- DOI: https://doi.org/10.1038/s41477-023-01421-0
- Primary Citation of Related Structures:
8BB6 - PubMed Abstract:
Sucrose import from photosynthetic tissues into the phloem is mediated by transporters from the low-affinity sucrose transporter family (SUC/SUT family). Furthermore, sucrose redistribution to other tissues is driven by phloem sap movement, the product of high turgor pressure created by this import activity. Additionally, sink organs such as fruits, cereals and seeds that accumulate high concentrations of sugar also depend on this active transport of sucrose. Here we present the structure of the sucrose-proton symporter, Arabidopsis thaliana SUC1, in an outward open conformation at 2.7 Å resolution, together with molecular dynamics simulations and biochemical characterization. We identify the key acidic residue required for proton-driven sucrose uptake and describe how protonation and sucrose binding are strongly coupled. Sucrose binding is a two-step process, with initial recognition mediated by the glucosyl moiety binding directly to the key acidic residue in a stringent pH-dependent manner. Our results explain how low-affinity sucrose transport is achieved in plants, and pinpoint a range of SUC binders that help define selectivity. Our data demonstrate a new mode for proton-driven symport with links to cation-driven symport and provide a broad model for general low-affinity transport in highly enriched substrate environments.
Organizational Affiliation:
Department of Molecular Biology and Genetics, Aarhus University, Aarhus, Denmark.