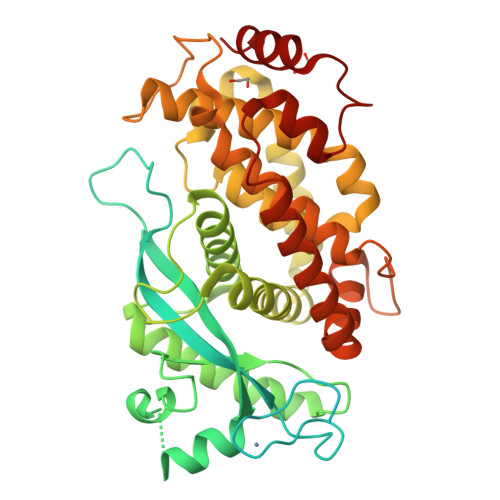
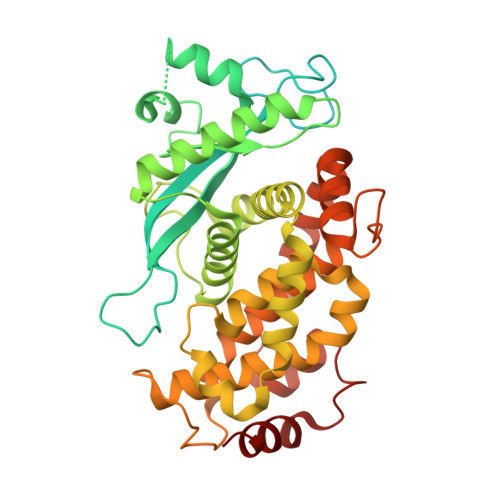
UBE2A and UBE2B are recruited by an atypical E3 ligase module in UBR4.
Barnsby-Greer, L., Mabbitt, P.D., Dery, M.A., Squair, D.R., Wood, N.T., Lamoliatte, F., Lange, S.M., Virdee, S.(2024) Nat Struct Mol Biol 31: 351-363
- PubMed: 38182926
- DOI: https://doi.org/10.1038/s41594-023-01192-4
- Primary Citation of Related Structures:
8B5W, 8BTL - PubMed Abstract:
UBR4 is a 574 kDa E3 ligase (E3) of the N-degron pathway with roles in neurodevelopment, age-associated muscular atrophy and cancer. The catalytic module that carries out ubiquitin (Ub) transfer remains unknown. Here we identify and characterize a distinct E3 module within human UBR4 consisting of a 'hemiRING' zinc finger, a helical-rich UBR zinc-finger interacting (UZI) subdomain, and an N-terminal region that can serve as an affinity factor for the E2 conjugating enzyme (E2). The structure of an E2-E3 complex provides atomic-level insight into the specificity determinants of the hemiRING toward the cognate E2s UBE2A/UBE2B. Via an allosteric mechanism, the UZI subdomain modestly activates the Ub-loaded E2 (E2∼Ub). We propose attenuated activation is complemented by the intrinsically high lysine reactivity of UBE2A, and their cooperation imparts a reactivity profile important for substrate specificity and optimal degradation kinetics. These findings reveal the mechanistic underpinnings of a neuronal N-degron E3, its specific recruitment of UBE2A, and highlight the underappreciated architectural diversity of cross-brace domains with Ub E3 activity.
Organizational Affiliation:
MRC Protein Phosphorylation and Ubiquitylation Unit, University of Dundee, Scotland, UK.