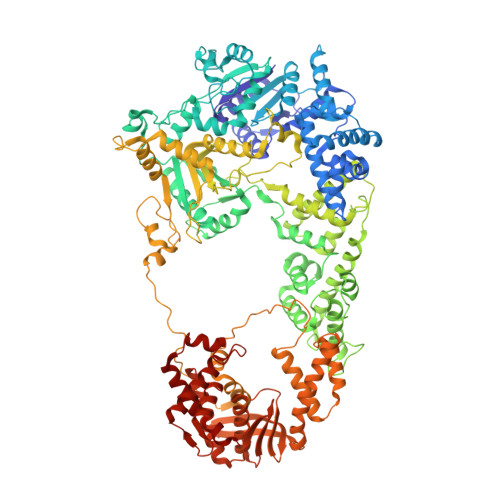
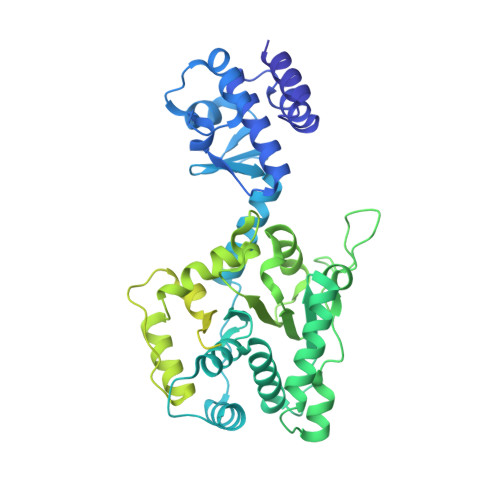
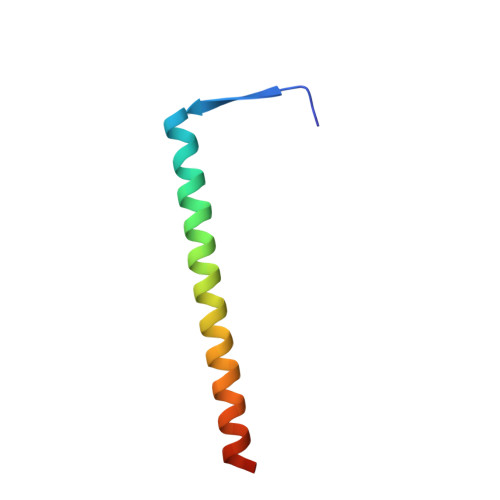
Structures of RecBCD in complex with phage-encoded inhibitor proteins reveal distinctive strategies for evasion of a bacterial immunity hub.
Wilkinson, M., Wilkinson, O.J., Feyerherm, C., Fletcher, E.E., Wigley, D.B., Dillingham, M.S.(2022) Elife 11
- PubMed: 36533901
- DOI: https://doi.org/10.7554/eLife.83409
- Primary Citation of Related Structures:
8B1R, 8B1T, 8B1U - PubMed Abstract:
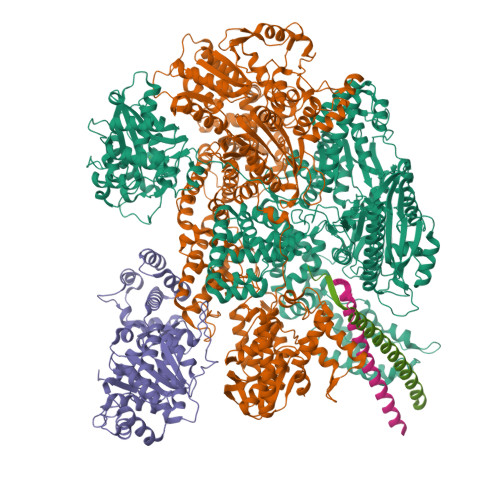
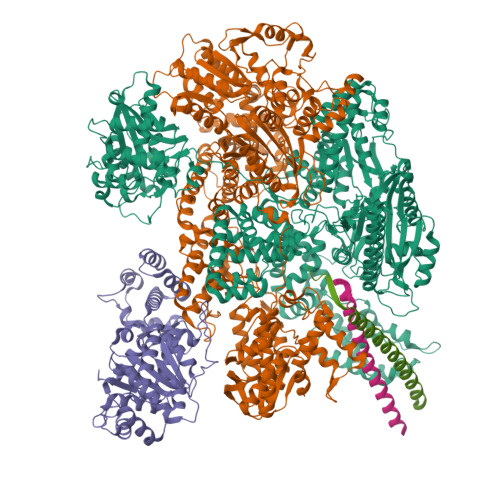
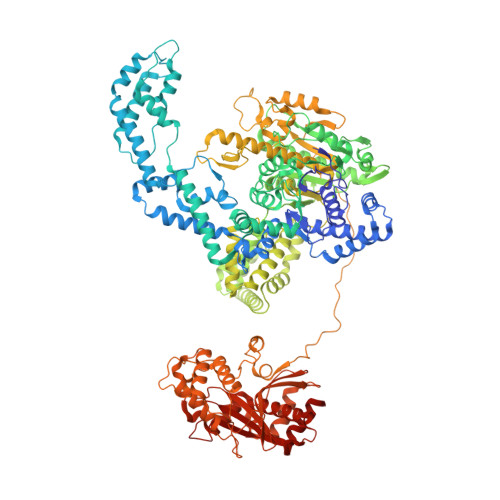
Following infection of bacterial cells, bacteriophage modulate double-stranded DNA break repair pathways to protect themselves from host immunity systems and prioritise their own recombinases. Here, we present biochemical and structural analysis of two phage proteins, gp5.9 and Abc2, which target the DNA break resection complex RecBCD. These exemplify two contrasting mechanisms for control of DNA break repair in which the RecBCD complex is either inhibited or co-opted for the benefit of the invading phage. Gp5.9 completely inhibits RecBCD by preventing it from binding to DNA. The RecBCD-gp5.9 structure shows that gp5.9 acts by substrate mimicry, binding predominantly to the RecB arm domain and competing sterically for the DNA binding site. Gp5.9 adopts a parallel coiled-coil architecture that is unprecedented for a natural DNA mimic protein. In contrast, binding of Abc2 does not substantially affect the biochemical activities of isolated RecBCD. The RecBCD-Abc2 structure shows that Abc2 binds to the Chi-recognition domains of the RecC subunit in a position that might enable it to mediate the loading of phage recombinases onto its single-stranded DNA products.
Organizational Affiliation:
Section of Structural Biology, Department of Infectious Disease, Faculty of Medicine, Imperial College London, London, United Kingdom.