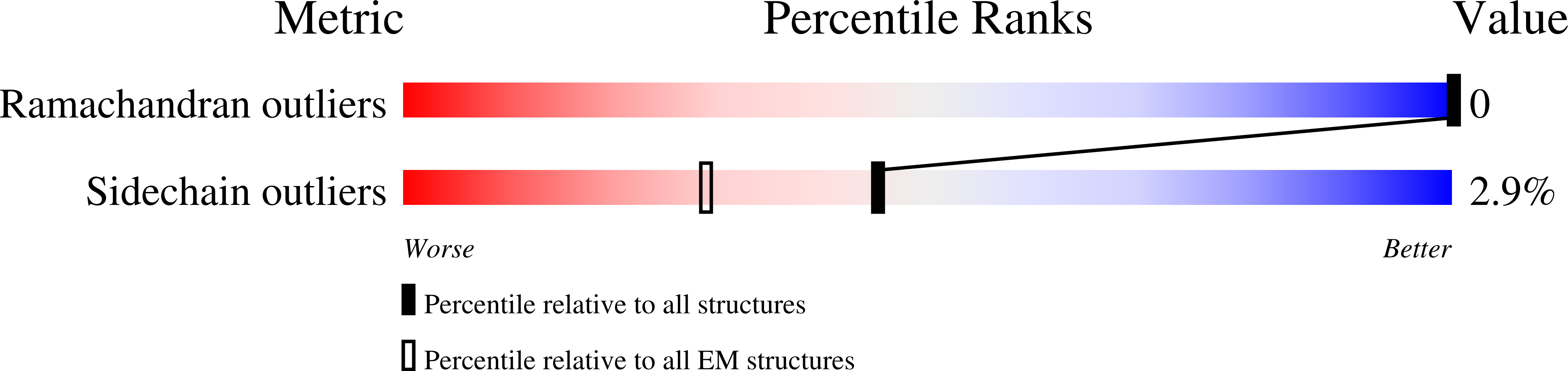Integrative structural analysis of the type III secretion system needle complex from Shigella flexneri.
Flacht, L., Lunelli, M., Kaszuba, K., Chen, Z.A., Reilly, F.J.O., Rappsilber, J., Kosinski, J., Kolbe, M.(2023) Protein Sci 32: e4595-e4595
- PubMed: 36790757
- DOI: https://doi.org/10.1002/pro.4595
- Primary Citation of Related Structures:
8AXK, 8AXL, 8AXN - PubMed Abstract:
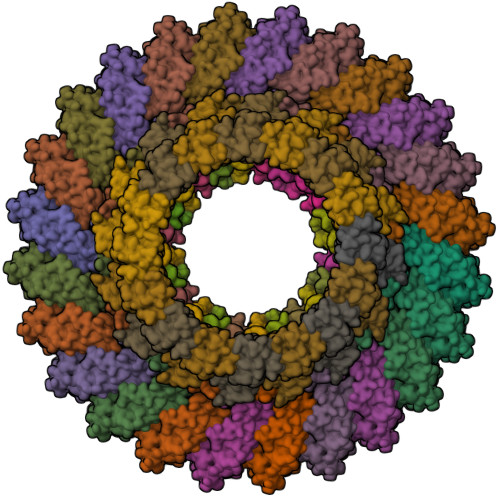
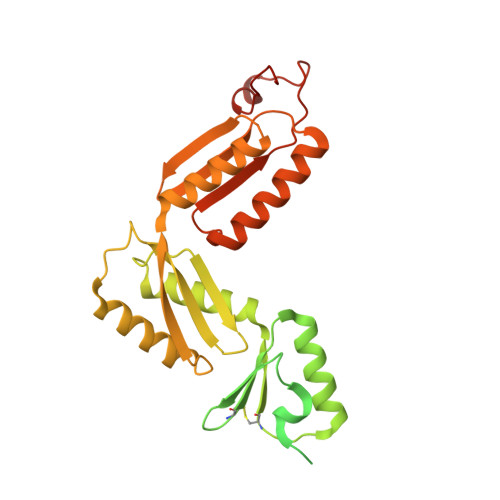
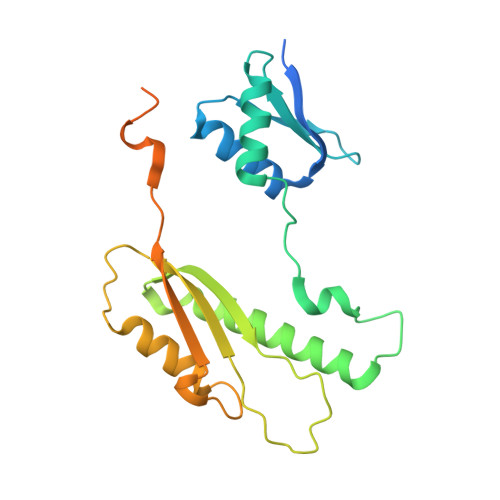
The type III secretion system (T3SS) is a large, transmembrane protein machinery used by various pathogenic gram-negative bacteria to transport virulence factors into the host cell during infection. Understanding the structure of T3SSs is crucial for future developments of therapeutics that could target this system. However, much of the knowledge about the structure of T3SS is available only for Salmonella, and it is unclear how this large assembly is conserved across species. Here, we combined cryo-electron microscopy, cross-linking mass spectrometry, and integrative modeling to determine the structure of the T3SS needle complex from Shigella flexneri. We show that the Shigella T3SS exhibits unique features distinguishing it from other structurally characterized T3SSs. The secretin pore complex adopts a new fold of its C-terminal S domain and the pilotin MxiM[SctG] locates around the outer surface of the pore. The export apparatus structure exhibits a conserved pseudohelical arrangement but includes the N-terminal domain of the SpaS[SctU] subunit, which was not present in any of the previously published virulence-related T3SS structures. Similar to other T3SSs, however, the apparatus is anchored within the needle complex by a network of flexible linkers that either adjust conformation to connect to equivalent patches on the secretin oligomer or bind distinct surface patches at the same height of the export apparatus. The conserved and unique features delineated by our analysis highlight the necessity to analyze T3SS in a species-specific manner, in order to fully understand the underlying molecular mechanisms of these systems. The structure of the type III secretion system from Shigella flexneri delineates conserved and unique features, which could be used for the development of broad-range therapeutics.
Organizational Affiliation:
Department for Structural Infection Biology, Center for Structural Systems Biology (CSSB) & Helmholtz Centre for Infection Research (HZI), Hamburg, Germany.