Oxazolo[5,4-f]quinoxaline-type selective inhibitors of glycogen synthase kinase-3 alpha (GSK-3 alpha ): Development and impact on temozolomide treatment of glioblastoma cells.
Hasyeoui, M., Lassagne, F., Erb, W., Nael, M., Elokely, K.M., Chaikuad, A., Knapp, S., Jorda, A., Valles, S.L., Quissac, E., Verreault, M., Robert, T., Bach, S., Samarat, A., Mongin, F.(2023) Bioorg Chem 134: 106456-106456
- PubMed: 36913879
- DOI: https://doi.org/10.1016/j.bioorg.2023.106456
- Primary Citation of Related Structures:
8AUZ, 8AV1 - PubMed Abstract:
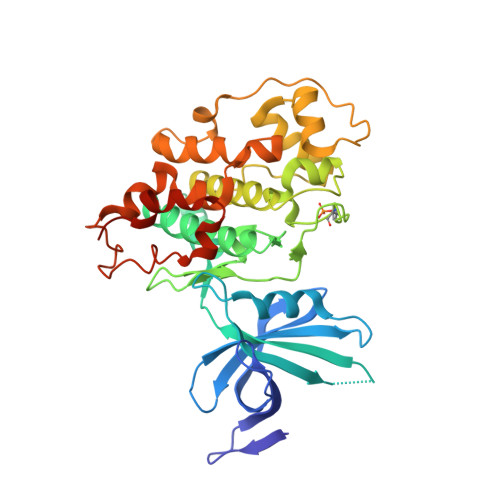
The 2-(3-pyridyl)oxazolo[5,4-f]quinoxalines CD-07 and FL-291 are ATP-competitive GSK-3 kinase inhibitors. Here, we investigated the impact of FL-291 on neuroblastoma cell viability and showed that treatment at 10 μM (i.e. ∼500 times the IC 50 against the GSK-3 isoforms) has no significant effect on the viability of NSC-34 motoneuron-like cells. A study performed on primary neurons (non-cancer cells) led to similar results. The structures co-crystallized with GSK-3β revealed similar binding modes for FL-291 and CD-07, with their hinge-oriented planar tricyclic system. Both GSK isoforms show the same orientations for the amino acids at the binding pocket except for Phe130 (α) and Phe67 (β), leading to a larger pocket on the opposite side of the hinge region for the α isoform. Calculations of the thermodynamic properties of the binding pockets highlighted the required features of potential ligands; these should have a hydrophobic core (which could be larger in the case of GSK-3β) surrounded by polar areas (a little more polar in the case of GSK-3α). A library of 27 analogs of FL-291 and CD-07 was thus designed and synthesized by taking advantage of this hypothesis. While the introduction of substituents at different positions of the pyridine ring, the replacement of the pyridine by other heterocyclic moieties, or the replacement of the quinoxaline ring by a quinoline moiety did not lead to any improvement, the replacement of the N-(thio)morpholino of FL-291/CD-07 by a slightly more polar N-thiazolidino led to a significant result. Indeed, the new inhibitor MH-124 showed clear selectivity for the α isoform, with IC 50 values of 17 nM and 239 nM on GSK-3α and GSK-3β, respectively. Finally, the efficacy of MH-124 was evaluated on two glioblastoma cell lines. Although MH-124 alone did not have a significant impact on cell survival, its addition to temozolomide (TMZ) significantly reduced the TMZ IC 50 values on the cells tested. The use of the Bliss model allowed a synergy to be evidenced at certain concentrations.
Organizational Affiliation:
Univ Rennes, CNRS, ISCR (Institut des Sciences Chimiques de Rennes) - UMR 6226, F-35000 Rennes, France; University of Carthage, Faculty of Sciences of Bizerte, LR18ES11, Laboratory of Hetero-Organic Compounds and Nanostructured Materials, 7021 Bizerte, Tunisia.