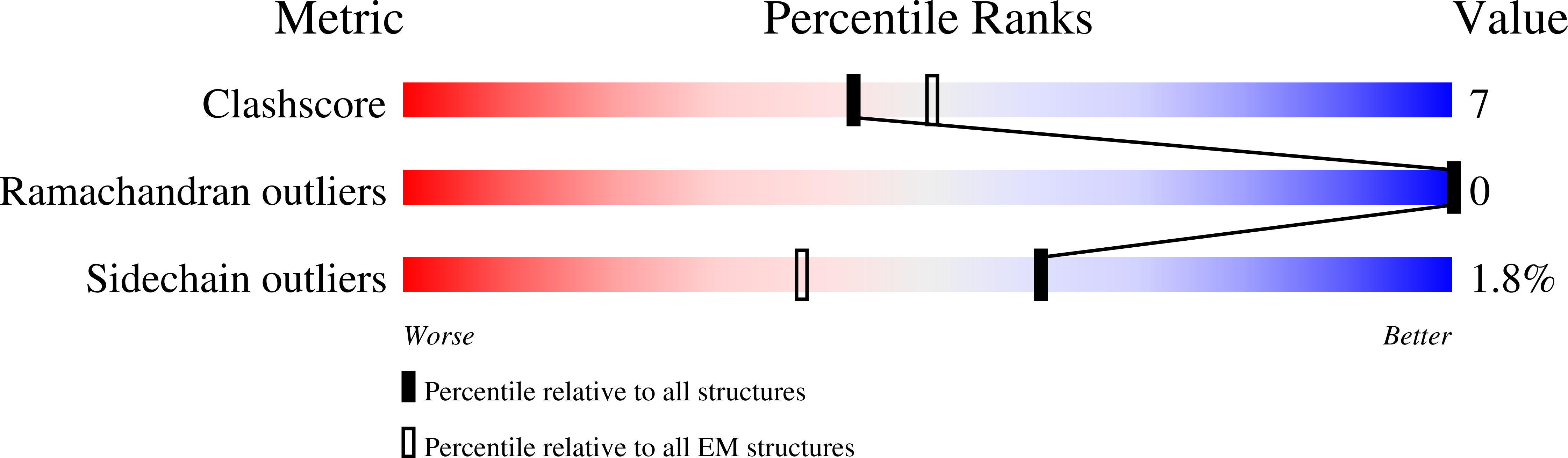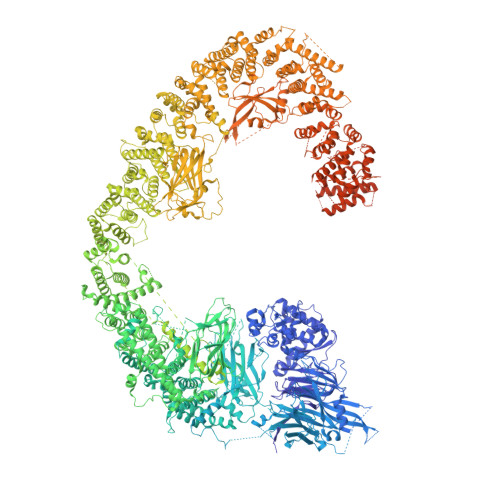Structural basis for regulation of apoptosis and autophagy by the BIRC6/SMAC complex.
Ehrmann, J.F., Grabarczyk, D.B., Heinke, M., Deszcz, L., Kurzbauer, R., Hudecz, O., Shulkina, A., Gogova, R., Meinhart, A., Versteeg, G.A., Clausen, T.(2023) Science 379: 1117-1123
- PubMed: 36758105
- DOI: https://doi.org/10.1126/science.ade8873
- Primary Citation of Related Structures:
8ATU, 8ATX, 8AUK, 8AUW - PubMed Abstract:
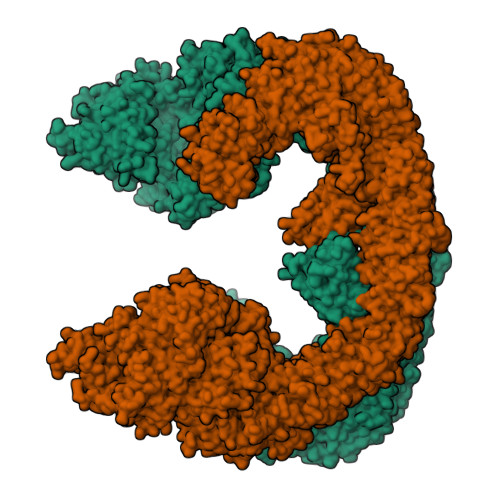
Inhibitor of apoptosis proteins (IAPs) bind to pro-apoptotic proteases, keeping them inactive and preventing cell death. The atypical ubiquitin ligase BIRC6 is the only essential IAP, additionally functioning as a suppressor of autophagy. We performed a structure-function analysis of BIRC6 in complex with caspase-9, HTRA2, SMAC, and LC3B, which are critical apoptosis and autophagy proteins. Cryo-electron microscopy structures showed that BIRC6 forms a megadalton crescent shape that arcs around a spacious cavity containing receptor sites for client proteins. Multivalent binding of SMAC obstructs client binding, impeding ubiquitination of both autophagy and apoptotic substrates. On the basis of these data, we discuss how the BIRC6/SMAC complex can act as a stress-induced hub to regulate apoptosis and autophagy drivers.
Organizational Affiliation:
Research Institute of Molecular Pathology, Vienna BioCenter, Vienna, Austria.